

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147014.14 + phase: 0 /pseudo
(342 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
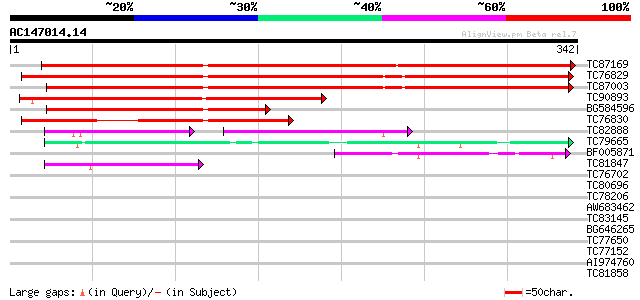
Score E
Sequences producing significant alignments: (bits) Value
TC87169 similar to PIR|G96689|G96689 probable fructokinase F28G1... 405 e-113
TC76829 similar to PIR|T14544|T14544 fructokinase (EC 2.7.1.4) -... 353 5e-98
TC87003 similar to PIR|T14544|T14544 fructokinase (EC 2.7.1.4) -... 349 1e-96
TC90893 similar to GP|23476263|gb|AAM44084.1 fructokinase {Lycop... 209 2e-54
BG584596 similar to GP|23476263|gb fructokinase {Lycopersicon es... 179 1e-45
TC76830 homologue to SP|P37829|SCRK_SOLTU Fructokinase (EC 2.7.1... 141 3e-34
TC82888 similar to PIR|A96716|A96716 probable fructokinase F23O1... 74 3e-22
TC79665 similar to PIR|F86307|F86307 hypothetical protein AAD500... 60 2e-09
BF005871 weakly similar to GP|20219054|gb Putative sugar kinase ... 49 4e-06
TC81847 weakly similar to GP|16226361|gb|AAL16146.1 unknown prot... 45 5e-05
TC76702 similar to SP|Q9LZG0|ADK2_ARATH Adenosine kinase 2 (EC 2... 40 0.002
TC80696 similar to GP|13605673|gb|AAK32830.1 At1g06730/F4H5_22 {... 39 0.004
TC78206 similar to GP|15450507|gb|AAK96546.1 AT4g27600/T29A15_90... 38 0.005
AW683462 homologue to GP|10177882|dbj fructokinase 1 {Arabidopsi... 38 0.006
TC83145 weakly similar to GP|16604509|gb|AAL24260.1 AT5g58730/mz... 35 0.042
BG646265 similar to SP|Q9SF85|ADK1 Adenosine kinase 1 (EC 2.7.1.... 35 0.055
TC77650 similar to GP|9795587|gb|AAF98405.1| Unknown protein {Ar... 34 0.071
TC77152 homologue to SP|P12859|G3PB_PEA Glyceraldehyde 3-phospha... 32 0.46
AI974760 similar to GP|16226361|gb| unknown protein {Arabidopsis... 31 0.79
TC81858 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 30 1.8
>TC87169 similar to PIR|G96689|G96689 probable fructokinase F28G11.11
[imported] - Arabidopsis thaliana, partial (84%)
Length = 1529
Score = 405 bits (1041), Expect = e-113
Identities = 204/322 (63%), Positives = 252/322 (77%)
Frame = +2
Query: 20 KGPLVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGN 79
+ PLVVCFGEM+I+ VPT+ G+SL+DA A+KK+P GA A V+V ISRLGGSSAFIGKVG
Sbjct: 380 ESPLVVCFGEMLIDFVPTVSGLSLADAPAFKKAPGGAPANVAVGISRLGGSSAFIGKVGE 559
Query: 80 DEFGHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPSADILFRS 139
DEFG+ML+DILK+N V++ G+ FD ARTALAF +L++ DG+ EFMFYRNPSAD+L +
Sbjct: 560 DEFGYMLADILKENNVNSQGMRFDPGARTALAFVTLRS--DGEREFMFYRNPSADMLLQE 733
Query: 140 EEIDKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAPNLTVPLWPSTEA 199
+E+D LI KA IFHYGS+SLI EP +S HI A AK G LSY PNL +PLWPS ++
Sbjct: 734 DELDLDLITKAKIFHYGSISLITEPCKSAHIAAAKAAKEAGVFLSYDPNLRLPLWPSADS 913
Query: 200 AREGIMSIWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGCRY 259
ARE I+SIW AD+IK+S EEI LT G DPYDD +++KLFH NLKLLLVTEG +GCRY
Sbjct: 914 ARERILSIWETADIIKISEEEISFLTNGEDPYDD-AVVRKLFHPNLKLLLVTEGAEGCRY 1090
Query: 260 YTKDFKGWVYGFEVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAA 319
YTK+F G V G +V+A+DTTGAGD+FV G LS L+ + + E+ LRE+L FANACGA
Sbjct: 1091YTKEFSGRVTGMKVDAVDTTGAGDAFVAGILSQLATDLSLLQKEEQLRESLRFANACGAL 1270
Query: 320 TVTGRGAIPSLPTKSSVLRVML 341
TVT RGAIP+LPTK +VL +L
Sbjct: 1271TVTERGAIPALPTKETVLNALL 1336
>TC76829 similar to PIR|T14544|T14544 fructokinase (EC 2.7.1.4) - beet,
partial (95%)
Length = 1408
Score = 353 bits (906), Expect = 5e-98
Identities = 184/333 (55%), Positives = 240/333 (71%)
Frame = +3
Query: 8 FIGSSKHGRKICKGPLVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRL 67
F +S +G L+V FGEM+I+ VPT+ GVSL++A + K+P GA A V++A+SRL
Sbjct: 63 FSMASSNGIPTTGTGLIVSFGEMLIDFVPTVSGVSLAEAPGFLKAPGGAPANVAIAVSRL 242
Query: 68 GGSSAFIGKVGNDEFGHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMF 127
GG +AF+GK+G+DEFGHML+ ILK+NGV G+ FD+ ARTALAF +L+ DG+ EFMF
Sbjct: 243 GGKAAFVGKLGDDEFGHMLAGILKENGVVAEGITFDQGARTALAFVTLRA--DGEREFMF 416
Query: 128 YRNPSADILFRSEEIDKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAP 187
YRNPSAD+L + EE++ LI+ A +FHYGS+SLI EP RS H++A+ AK G +LSY P
Sbjct: 417 YRNPSADMLLKPEELNLELIRSAKVFHYGSISLIVEPCRSAHLKALEVAKEAGCLLSYDP 596
Query: 188 NLTVPLWPSTEAAREGIMSIWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKL 247
NL +PLWPS + AR+ I+SIW AD+IKVS E+ LT G+D DD + L+H NLKL
Sbjct: 597 NLRLPLWPSADEARKQILSIWEKADLIKVSDNELEFLT-GSDKIDDATAL-TLWHPNLKL 770
Query: 248 LLVTEGIKGCRYYTKDFKGWVYGFEVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILR 307
LLVT G G RYYTK+F G V GF+V +DTTGAGDSFVG L+ + + I +DE LR
Sbjct: 771 LLVTLGEHGARYYTKNFHGQVDGFKVNTVDTTGAGDSFVGALLAKIVDDQAILEDESRLR 950
Query: 308 EALDFANACGAATVTGRGAIPSLPTKSSVLRVM 340
E L FANACGA T T +GAIP+LP + VL ++
Sbjct: 951 EVLKFANACGAITTTKKGAIPALPKEEDVLNLI 1049
>TC87003 similar to PIR|T14544|T14544 fructokinase (EC 2.7.1.4) - beet,
partial (95%)
Length = 1305
Score = 349 bits (895), Expect = 1e-96
Identities = 182/318 (57%), Positives = 233/318 (73%)
Frame = +3
Query: 23 LVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGNDEF 82
L+V FGEM+I+ VPT GVSL++A + K+P GA A V++A+SRLGG SAF+GK+G+DEF
Sbjct: 81 LIVSFGEMLIDFVPTASGVSLAEAPGFLKAPGGAPANVAIAVSRLGGKSAFVGKLGDDEF 260
Query: 83 GHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPSADILFRSEEI 142
GHML+ ILK+N V G+ FD+ ARTALAF +L+ DG+ EFMFYRNPSAD+L E++
Sbjct: 261 GHMLAGILKENNVRGDGINFDKGARTALAFVTLRA--DGEREFMFYRNPSADMLLTPEDL 434
Query: 143 DKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAPNLTVPLWPSTEAARE 202
+ LI+ A +FHYGS+SLI EP RS H++A+ AK G +LSY PNL +PLWPS E AR+
Sbjct: 435 NLELIRSAKVFHYGSISLIVEPCRSAHLKAMEVAKDAGCLLSYDPNLRLPLWPSPEEARK 614
Query: 203 GIMSIWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGCRYYTK 262
I+SIW+ AD+IKVS E+ LT G+D DD + L+H NLKLLLVT G G RYYTK
Sbjct: 615 QILSIWDKADLIKVSDVELEFLT-GSDKIDDASAL-SLWHPNLKLLLVTLGENGSRYYTK 788
Query: 263 DFKGWVYGFEVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAATVT 322
+FKG V F V +DTTGAGDSFVG L + + I +DE LRE L FANACGA T T
Sbjct: 789 NFKGSVDAFHVNTVDTTGAGDSFVGALLGKIVDDQSILEDEARLREVLKFANACGAITTT 968
Query: 323 GRGAIPSLPTKSSVLRVM 340
+GAIP+LPT++ VL ++
Sbjct: 969 KKGAIPALPTEADVLSLI 1022
>TC90893 similar to GP|23476263|gb|AAM44084.1 fructokinase {Lycopersicon
esculentum}, partial (43%)
Length = 664
Score = 209 bits (531), Expect = 2e-54
Identities = 112/198 (56%), Positives = 141/198 (70%), Gaps = 13/198 (6%)
Frame = +2
Query: 7 LFIGSS------------KHGRKIC-KGPLVVCFGEMMINLVPTIDGVSLSDAEAYKKSP 53
LF GSS H R++ K LVVCFGE++I+ VPT+ GVSL++A A+KK+P
Sbjct: 59 LFFGSSDGTVYPLR*I**SHKRRVQPKVDLVVCFGELLIDFVPTVGGVSLAEAPAFKKAP 238
Query: 54 AGATAIVSVAISRLGGSSAFIGKVGNDEFGHMLSDILKQNGVDNSGLLFDEHARTALAFH 113
GA A V+V ISRLGGSSAF+GKVG DEFG+ML+DILKQN VD SG+ FD ARTALAF
Sbjct: 239 GGAPANVAVGISRLGGSSAFLGKVGADEFGYMLADILKQNNVDTSGMRFDSDARTALAFV 418
Query: 114 SLKNSDDGKPEFMFYRNPSADILFRSEEIDKSLIKKATIFHYGSVSLIKEPSRSTHIEAI 173
+L+ DG+ EF+F+RNP AD+L E+D +LI+KA IFHYGS+SLI EP +S H+ A+
Sbjct: 419 TLR--ADGEREFLFFRNPXADMLLDKSELDHNLIEKAKIFHYGSISLIDEPCKSAHLAAL 592
Query: 174 NYAKMCGSILSYAPNLTV 191
AK ILSY PN +
Sbjct: 593 RIAKDSDCILSYDPNFXI 646
>BG584596 similar to GP|23476263|gb fructokinase {Lycopersicon esculentum},
partial (35%)
Length = 758
Score = 179 bits (455), Expect = 1e-45
Identities = 91/135 (67%), Positives = 111/135 (81%)
Frame = +2
Query: 23 LVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGNDEF 82
LVV FGEM+I+ VPT+ GVSL++A A+KKSP GA A V+V ISRLGGSSAFIGKVG DEF
Sbjct: 359 LVVSFGEMLIDFVPTVGGVSLAEAPAFKKSPGGAPANVAVGISRLGGSSAFIGKVGADEF 538
Query: 83 GHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPSADILFRSEEI 142
G+ML+DILKQN VD SG+ FD +ARTALAF +L++ DG+ EF+F+RNPSAD+L E+
Sbjct: 539 GYMLADILKQNNVDTSGMRFDSNARTALAFVTLRS--DGEREFLFFRNPSADMLLHESEL 712
Query: 143 DKSLIKKATIFHYGS 157
D L+KKA IFHYGS
Sbjct: 713 DIDLLKKARIFHYGS 757
>TC76830 homologue to SP|P37829|SCRK_SOLTU Fructokinase (EC 2.7.1.4).
[Potato] {Solanum tuberosum}, partial (38%)
Length = 436
Score = 141 bits (356), Expect = 3e-34
Identities = 77/164 (46%), Positives = 105/164 (63%)
Frame = +3
Query: 8 FIGSSKHGRKICKGPLVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRL 67
F +S +G L+V FGEM+I+ VPT+ GVSL++A ++ K+P
Sbjct: 21 FSMASSNGIPTTGTGLIVSFGEMLIDFVPTVSGVSLAEAPSFLKAP-------------- 158
Query: 68 GGSSAFIGKVGNDEFGHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMF 127
G+DEFGHML+ ILK+NGV G+ FD+ ARTALAF +L+ DG+ EFMF
Sbjct: 159 ----------GDDEFGHMLAGILKENGVVAEGITFDQGARTALAFVTLRA--DGEREFMF 302
Query: 128 YRNPSADILFRSEEIDKSLIKKATIFHYGSVSLIKEPSRSTHIE 171
YRNPSAD+L + EE++ LI+ A +FHYGS+SLI EP RS H++
Sbjct: 303 YRNPSADMLLKPEELNLELIRSAKVFHYGSISLIVEPCRSAHLK 434
>TC82888 similar to PIR|A96716|A96716 probable fructokinase F23O10.21
[imported] - Arabidopsis thaliana, partial (42%)
Length = 1053
Score = 73.6 bits (179), Expect(2) = 3e-22
Identities = 39/116 (33%), Positives = 64/116 (54%), Gaps = 2/116 (1%)
Frame = +3
Query: 130 NPSADILFRSEEIDKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAPNL 189
NP I EI+ ++K+A +F++ + SL+ RST + AI AK G+++ Y NL
Sbjct: 621 NPVLKIXLTKSEINIDVLKEAKMFYFNTHSLLDRHMRSTTLRAIKIAKHFGAVVFYDVNL 800
Query: 190 TVPLWPSTEAAREGIMSIWNYADVIKVSVEEIRIL--TEGNDPYDDKMIMKKLFHH 243
+PLW S E + I +WN AD+I+V+ +E+ L ++ +D K + F H
Sbjct: 801 PMPLWHSQEETKTFIQQVWNLADIIEVTKQELEFLCGITPSEEFDTKNNARSKFVH 968
Score = 49.3 bits (116), Expect(2) = 3e-22
Identities = 30/104 (28%), Positives = 48/104 (45%), Gaps = 14/104 (13%)
Frame = +2
Query: 22 PLVVCFGEMMINLVPT-------IDGV-------SLSDAEAYKKSPAGATAIVSVAISRL 67
PLV CFG VP+ ID +L E + ++P G V++A++ L
Sbjct: 266 PLVCCFGAAQHAFVPSGRPANRLIDHELHERMKDALWSPEKFVRAPGGCAGSVAIALASL 445
Query: 68 GGSSAFIGKVGNDEFGHMLSDILKQNGVDNSGLLFDEHARTALA 111
GG AF+GK+ +DE+G + + N V + D T ++
Sbjct: 446 GGKVAFMGKLADDEYGQAMLYYMNANNVQTRSVSIDSKRATGVS 577
>TC79665 similar to PIR|F86307|F86307 hypothetical protein AAD50017.1
[imported] - Arabidopsis thaliana, partial (78%)
Length = 1402
Score = 59.7 bits (143), Expect = 2e-09
Identities = 82/329 (24%), Positives = 134/329 (39%), Gaps = 10/329 (3%)
Frame = +1
Query: 22 PLVVCFGEMMINLVPTID-----GVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGK 76
P VV G ++ ID G ++S A++ + G A + ++L + F+G+
Sbjct: 151 PPVVVVGSANADIYVEIDRLPKEGETIS-AKSGQTLAGGKGANQACCGAKLSHPTYFLGQ 327
Query: 77 VGNDEFGHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFYRNPSADIL 136
VGND G++++D L+ GV L A T A L++S + N S
Sbjct: 328 VGNDAHGNLVADALRDGGVRLDYLAVVSSAPTGHAVVMLQSSGQNSIVIVGGANVSC--- 498
Query: 137 FRSEEIDKSLIKKATIFHYGSVSLIKEPSRSTHIEAINYAKMCG-SILSYAPNLTVPLWP 195
+ K L + G V L +E +++ AK G ++ A + P+
Sbjct: 499 WPQTLPPKQL---EVVSSAGIVLLQREIPDFVNVQVAKAAKSAGVPVIFDAGGMDAPI-- 663
Query: 196 STEAAREGIMSIWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNL--KLLLVTEG 253
+ ++ D+ + E+ LT +++ H L K +LV G
Sbjct: 664 --------PQELLDFVDIFSPNESELGRLTGLPTESFEEITQAAAKCHKLGVKQVLVKLG 819
Query: 254 IKGCRYYTKDFKGWVYG--FEVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALD 311
KG + + F IDTTGAGD+F F L + K +E L
Sbjct: 820 AKGSALFIDGEEPIQQPAIFAKTVIDTTGAGDTFTAAFAVALV-------EGKSKKECLR 978
Query: 312 FANACGAATVTGRGAIPSLPTKSSVLRVM 340
FA A + V +GAIPS+P + SVL ++
Sbjct: 979 FAAAAASLCVQVKGAIPSMPDRKSVLELL 1065
>BF005871 weakly similar to GP|20219054|gb Putative sugar kinase {Oryza
sativa (japonica cultivar-group)}, partial (33%)
Length = 563
Score = 48.5 bits (114), Expect = 4e-06
Identities = 43/147 (29%), Positives = 66/147 (44%), Gaps = 5/147 (3%)
Frame = +2
Query: 197 TEAAREGIMSIWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNL--KLLLVTEGI 254
T+ + + +DV+ ++ +E LT DP + ++L + K ++V G+
Sbjct: 38 TQEQQRALNQFLRMSDVLLLTSDEAESLTGIGDPI---LAGQELLKRGIRTKWVIVKMGL 208
Query: 255 KGCRYYTKDFKGWVYGFEVEAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFAN 314
KG T F+V +D+ G GDSFV A+ I+K + L FAN
Sbjct: 209 KGSILITSSSIACAPAFKVNIVDSVGCGDSFVAAI-----AYGFIHKLPMV--NTLAFAN 367
Query: 315 ACGAATVTGRGA---IPSLPTKSSVLR 338
A GAAT G GA + SL +LR
Sbjct: 368 AVGAATAMGCGAGRNVTSLEKVVDILR 448
>TC81847 weakly similar to GP|16226361|gb|AAL16146.1 unknown protein
{Arabidopsis thaliana}, partial (29%)
Length = 762
Score = 44.7 bits (104), Expect = 5e-05
Identities = 27/100 (27%), Positives = 46/100 (46%), Gaps = 4/100 (4%)
Frame = +2
Query: 22 PLVVCFGEMMINLVPTIDGVSLSDAE----AYKKSPAGATAIVSVAISRLGGSSAFIGKV 77
P++V FG + ++L+ T++ D + +K G T +RLG I KV
Sbjct: 332 PVIVGFGGVGVDLLATVESFPKPDTKNRTTQFKVQGGGNTGNALTCAARLGLKPRIISKV 511
Query: 78 GNDEFGHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKN 117
ND G +L + L+ GVD S + + T ++ + N
Sbjct: 512 ANDAQGRVLIEELEAEGVDTSSFVVSKEGTTPFSYIIIDN 631
>TC76702 similar to SP|Q9LZG0|ADK2_ARATH Adenosine kinase 2 (EC 2.7.1.20)
(AK 2) (Adenosine 5'- phosphotransferase 2). [Mouse-ear
cress], partial (97%)
Length = 1592
Score = 39.7 bits (91), Expect = 0.002
Identities = 61/267 (22%), Positives = 100/267 (36%), Gaps = 36/267 (13%)
Frame = +1
Query: 69 GSSAFIGKVGNDEFGHMLSDILKQNGVDNSGLLFDEHARTALAFHSLKNSDDGKPEFMFY 128
G++++IG +G D+FG ++ KQ GV N DE+ T + E
Sbjct: 373 GATSYIGCIGKDKFGEEMTKNSKQAGV-NVHYYEDENTPTGTCAVCVVGG-----ERSLI 534
Query: 129 RNPSADILFRSEEIDK----SLIKKATIFHYGSVSLIKEPSR----STHIEAINYAKMCG 180
N SA ++ + + + +L++KA F+ L P + H A N M
Sbjct: 535 ANLSAANCYKVDHLKQPENWALVEKAKYFYIAGFFLTVSPESIQLVAEHAAANNKVFMMN 714
Query: 181 SILSYA------------PNLTVPLWPSTEAAREGIMSIWNYADVIKVSVE--------- 219
+ P + TEA + W +V +++++
Sbjct: 715 LSAPFICEFFKDPQEKALPYMDYVFGNETEARTFSKVHGWETENVEEIALKISQLPKASE 894
Query: 220 ---EIRILTEGNDPY----DDKMIMKKLFHHNLKLLLVTEGIKGCRYYTKDFKGWVYGFE 272
I ++T+G DP D K+ + + +LL E +
Sbjct: 895 ARKRITVITQGADPVCVAQDGKVTLYPV------ILLPKEKL------------------ 1002
Query: 273 VEAIDTTGAGDSFVGGFLSILSAHKHI 299
+DT GAGD+FVGGFLS L K I
Sbjct: 1003---VDTNGAGDAFVGGFLSQLVQEKPI 1074
>TC80696 similar to GP|13605673|gb|AAK32830.1 At1g06730/F4H5_22 {Arabidopsis
thaliana}, partial (39%)
Length = 1384
Score = 38.5 bits (88), Expect = 0.004
Identities = 17/47 (36%), Positives = 29/47 (61%)
Frame = +2
Query: 54 AGATAIVSVAISRLGGSSAFIGKVGNDEFGHMLSDILKQNGVDNSGL 100
AG +++A +RLG + IG VG + +G+ LSD+L+ G+ G+
Sbjct: 434 AGGNCNMAIAAARLGLNCVSIGHVGKEIYGNFLSDVLRDEGIGMVGM 574
>TC78206 similar to GP|15450507|gb|AAK96546.1 AT4g27600/T29A15_90 {Arabidopsis
thaliana}, partial (76%)
Length = 1677
Score = 38.1 bits (87), Expect = 0.005
Identities = 23/84 (27%), Positives = 41/84 (48%)
Frame = +1
Query: 207 IWNYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGCRYYTKDFKG 266
+ NYAD+I + +E R L D + + + + + L+ VT+G++G K
Sbjct: 1114 VGNYADLIFANADEARALCNF-DAKESTVSVTRYLSQFVPLVSVTDGLRGSYIGVKGEAV 1290
Query: 267 WVYGFEVEAIDTTGAGDSFVGGFL 290
++ +DT GAGD++ G L
Sbjct: 1291 YIPPSPCVPVDTCGAGDAYASGIL 1362
Score = 27.3 bits (59), Expect = 8.7
Identities = 18/59 (30%), Positives = 28/59 (46%), Gaps = 8/59 (13%)
Frame = +3
Query: 45 DAEAYKKSPAGATAIVSVAISRLGGSS--------AFIGKVGNDEFGHMLSDILKQNGV 95
D +YK + G+ + VA++RLGG S A G V +D G + L++ V
Sbjct: 627 DGCSYKAAAGGSLSNTLVALARLGGRSLRDPAINVAMAGSVASDLLGGFYREKLRRANV 803
>AW683462 homologue to GP|10177882|dbj fructokinase 1 {Arabidopsis thaliana},
partial (6%)
Length = 150
Score = 37.7 bits (86), Expect = 0.006
Identities = 15/23 (65%), Positives = 21/23 (91%)
Frame = +3
Query: 23 LVVCFGEMMINLVPTIDGVSLSD 45
LVVCFGE++I+ VPT+ GVSL++
Sbjct: 81 LVVCFGELLIDFVPTVGGVSLAE 149
>TC83145 weakly similar to GP|16604509|gb|AAL24260.1 AT5g58730/mzn1_180
{Arabidopsis thaliana}, partial (33%)
Length = 904
Score = 35.0 bits (79), Expect = 0.042
Identities = 18/41 (43%), Positives = 24/41 (57%)
Frame = +1
Query: 248 LLVTEGIKGCRYYTKDFKGWVYGFEVEAIDTTGAGDSFVGG 288
++VT G GC ++KD V FE +D TGA D F+GG
Sbjct: 766 VVVTHGKDGCEVFSKDGCLMVDPFESCQVDPTGARDCFLGG 888
>BG646265 similar to SP|Q9SF85|ADK1 Adenosine kinase 1 (EC 2.7.1.20) (AK 1)
(Adenosine 5'- phosphotransferase 1). [Mouse-ear cress],
partial (39%)
Length = 715
Score = 34.7 bits (78), Expect = 0.055
Identities = 16/24 (66%), Positives = 18/24 (74%)
Frame = -1
Query: 276 IDTTGAGDSFVGGFLSILSAHKHI 299
+DT GAGD+FVGGFLS L K I
Sbjct: 460 VDTNGAGDAFVGGFLSQLVQEKPI 389
>TC77650 similar to GP|9795587|gb|AAF98405.1| Unknown protein {Arabidopsis
thaliana}, partial (91%)
Length = 1530
Score = 34.3 bits (77), Expect = 0.071
Identities = 63/271 (23%), Positives = 110/271 (40%), Gaps = 9/271 (3%)
Frame = +1
Query: 29 EMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVA--ISRLGGSSAFIGKVGNDEFGHML 86
E + N++ + S S + K+ AG + ++ S G SS IG G+DE G +
Sbjct: 388 EELENILKEVKSKSNSADSSVMKTLAGGSVANTIRGLSSGFGISSGIIGACGDDEQGQLF 567
Query: 87 SDILKQNGVDNSGLLFDE-HARTALAFHSLKNSDDGKPEFMFYRNPSADILFRSEEIDKS 145
+ + NGVD S L + H + + +P S + +++E+
Sbjct: 568 VNNMSSNGVDLSRLRKKKGHTAQCVCLVDELGNRTMRPCL------SNAVKVQAQELMTE 729
Query: 146 LIK--KATIFHYGSVSLIKEPSRSTHIEAINYAKMCGSILSYAPNLTVPLWPSTEAAREG 203
K K + Y ++L AI AK G ++S L + + ++
Sbjct: 730 DFKGSKWLVLRYAILNL------EVIQAAIALAKQEGLLVS----LDLASFEMVRNFKQP 879
Query: 204 IMSIW--NYADVIKVSVEEIRILTEGNDPYDDKMIMKKLFHHNLKLLLVTEGIKGC--RY 259
++++ D+ + +E L G D ++ L + + +VT G GC R+
Sbjct: 880 LLNLLESGNVDLCFANEDEATELLRGEQNADPIAAVEFLAKY-CQWAVVTLGSNGCIARH 1056
Query: 260 YTKDFKGWVYGFEVEAIDTTGAGDSFVGGFL 290
+ + G E +A D TGAGD F GFL
Sbjct: 1057GKEMIRVSAIG-ESKATDATGAGDLFASGFL 1146
>TC77152 homologue to SP|P12859|G3PB_PEA Glyceraldehyde 3-phosphate
dehydrogenase B chloroplast precursor (EC 1.2.1.13),
complete
Length = 1782
Score = 31.6 bits (70), Expect = 0.46
Identities = 18/71 (25%), Positives = 36/71 (50%)
Frame = +3
Query: 20 KGPLVVCFGEMMINLVPTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSAFIGKVGN 79
+ PL + + E+ I++V GV + A K AGA ++ A ++ ++ V
Sbjct: 675 RDPLKLPWAELGIDIVIEGTGVFVDGPGAGKHIQAGAKKVIITAPAKGADIPTYVVGVNE 854
Query: 80 DEFGHMLSDIL 90
++GH ++DI+
Sbjct: 855 QDYGHEVADII 887
>AI974760 similar to GP|16226361|gb| unknown protein {Arabidopsis thaliana},
partial (19%)
Length = 432
Score = 30.8 bits (68), Expect = 0.79
Identities = 21/61 (34%), Positives = 31/61 (50%)
Frame = -2
Query: 274 EAIDTTGAGDSFVGGFLSILSAHKHIYKDEKILREALDFANACGAATVTGRGAIPSLPTK 333
E IDTTGAGD+F+G + + ++ +I+ L FA AA GA LP +
Sbjct: 374 ELIDTTGAGDAFIGAIMYAICSN----MAPEIM---LPFAAQVAAAKCRALGARTGLPHR 216
Query: 334 S 334
+
Sbjct: 215 T 213
>TC81858 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (9%)
Length = 748
Score = 29.6 bits (65), Expect = 1.8
Identities = 16/37 (43%), Positives = 24/37 (64%)
Frame = +1
Query: 36 PTIDGVSLSDAEAYKKSPAGATAIVSVAISRLGGSSA 72
P G ++SDA A ++PAG++A S +S +GGS A
Sbjct: 388 PVASGPAVSDAPA--EAPAGSSAAASFRVSFVGGSVA 492
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,504,932
Number of Sequences: 36976
Number of extensions: 139262
Number of successful extensions: 614
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 597
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 603
length of query: 342
length of database: 9,014,727
effective HSP length: 97
effective length of query: 245
effective length of database: 5,428,055
effective search space: 1329873475
effective search space used: 1329873475
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC147014.14


