

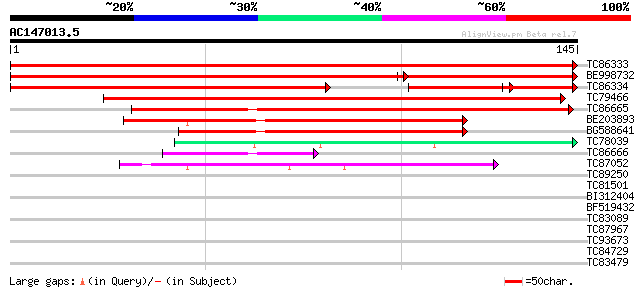
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147013.5 - phase: 0
(145 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86333 weakly similar to PIR|T00839|T00839 hypothetical protein... 292 3e-80
BE998732 weakly similar to PIR|T00839|T008 hypothetical protein ... 204 3e-75
TC86334 weakly similar to GP|10177132|dbj|BAB10422. senescence-a... 159 4e-57
TC79466 similar to PIR|T00839|T00839 hypothetical protein At2g17... 168 8e-43
TC86665 similar to GP|7594903|dbj|BAA88985.2 Ntdin {Nicotiana ta... 109 3e-25
BE203893 similar to GP|20197696|gb| senescence-associated protei... 89 6e-19
BG588641 similar to GP|20197696|gb| senescence-associated protei... 75 1e-14
TC78039 similar to PIR|T05875|T05875 hypothetical protein T29A15... 45 1e-05
TC86666 similar to GP|7594903|dbj|BAA88985.2 Ntdin {Nicotiana ta... 41 1e-04
TC87052 similar to GP|15010630|gb|AAK73974.1 At2g42220/T24P15.13... 40 3e-04
TC89250 weakly similar to GP|21592651|gb|AAM64600.1 rhodanese-li... 36 0.006
TC81501 27 2.2
BI312404 26 4.9
BF519432 similar to GP|6682233|gb| putative pectate lyase {Arabi... 26 4.9
TC83089 26 4.9
TC87967 homologue to GP|12720419|gb|AAK02280.1 unknown {Pasteure... 26 4.9
TC93673 similar to GP|16323198|gb|AAL15333.1 At1g15980/T24D18_8 ... 26 6.4
TC84729 homologue to PIR|C96615|C96615 hypothetical protein T18I... 26 6.4
TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protei... 25 8.4
>TC86333 weakly similar to PIR|T00839|T00839 hypothetical protein At2g17850
[imported] - Arabidopsis thaliana, partial (58%)
Length = 941
Score = 292 bits (748), Expect = 3e-80
Identities = 145/145 (100%), Positives = 145/145 (100%)
Frame = +1
Query: 1 MAAVSANLLPRCLAFFLLLVFVLCSSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKG 60
MAAVSANLLPRCLAFFLLLVFVLCSSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKG
Sbjct: 199 MAAVSANLLPRCLAFFLLLVFVLCSSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKG 378
Query: 61 HVDATKIINIPYLLDTPKGRVKNLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLAD 120
HVDATKIINIPYLLDTPKGRVKNLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLAD
Sbjct: 379 HVDATKIINIPYLLDTPKGRVKNLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLAD 558
Query: 121 GFKNVHNMGGGYMEWVSNKLPVIQQ 145
GFKNVHNMGGGYMEWVSNKLPVIQQ
Sbjct: 559 GFKNVHNMGGGYMEWVSNKLPVIQQ 633
>BE998732 weakly similar to PIR|T00839|T008 hypothetical protein At2g17850
[imported] - Arabidopsis thaliana, partial (30%)
Length = 709
Score = 204 bits (518), Expect(2) = 3e-75
Identities = 102/102 (100%), Positives = 102/102 (100%)
Frame = +1
Query: 1 MAAVSANLLPRCLAFFLLLVFVLCSSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKG 60
MAAVSANLLPRCLAFFLLLVFVLCSSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKG
Sbjct: 76 MAAVSANLLPRCLAFFLLLVFVLCSSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKG 255
Query: 61 HVDATKIINIPYLLDTPKGRVKNLNFVKQVSSSCDKEDCLVV 102
HVDATKIINIPYLLDTPKGRVKNLNFVKQVSSSCDKEDCLVV
Sbjct: 256 HVDATKIINIPYLLDTPKGRVKNLNFVKQVSSSCDKEDCLVV 381
Score = 94.0 bits (232), Expect(2) = 3e-75
Identities = 43/46 (93%), Positives = 45/46 (97%)
Frame = +2
Query: 100 LVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPVIQQ 145
+ +GCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPVIQQ
Sbjct: 395 IFLGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPVIQQ 532
>TC86334 weakly similar to GP|10177132|dbj|BAB10422. senescence-associated
protein sen1-like protein {Arabidopsis thaliana},
partial (53%)
Length = 593
Score = 159 bits (401), Expect(3) = 4e-57
Identities = 80/82 (97%), Positives = 80/82 (97%)
Frame = +1
Query: 1 MAAVSANLLPRCLAFFLLLVFVLCSSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKG 60
MAAVSANLLPRCLAFFLLLVFVLCSSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKG
Sbjct: 52 MAAVSANLLPRCLAFFLLLVFVLCSSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKG 231
Query: 61 HVDATKIINIPYLLDTPKGRVK 82
HVDATKIINIPYLLDTPKG K
Sbjct: 232 HVDATKIINIPYLLDTPKGLPK 297
Score = 54.3 bits (129), Expect(3) = 4e-57
Identities = 25/27 (92%), Positives = 25/27 (92%)
Frame = +2
Query: 103 GCQSGKRSFSATSELLADGFKNVHNMG 129
GCQSGKRSFSATSELLADGFKNVH G
Sbjct: 287 GCQSGKRSFSATSELLADGFKNVHQHG 367
Score = 45.1 bits (105), Expect(3) = 4e-57
Identities = 19/19 (100%), Positives = 19/19 (100%)
Frame = +3
Query: 127 NMGGGYMEWVSNKLPVIQQ 145
NMGGGYMEWVSNKLPVIQQ
Sbjct: 360 NMGGGYMEWVSNKLPVIQQ 416
>TC79466 similar to PIR|T00839|T00839 hypothetical protein At2g17850
[imported] - Arabidopsis thaliana, partial (56%)
Length = 652
Score = 168 bits (425), Expect = 8e-43
Identities = 79/118 (66%), Positives = 100/118 (83%)
Frame = +3
Query: 25 SSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNL 84
SS +VVT+DV A K+LI+T H+YLDVRTVEEF+KGHVD+ KIINI Y+ +TP+GRVKN
Sbjct: 75 SSKTEVVTVDVLATKSLIKTTHVYLDVRTVEEFQKGHVDSEKIINIAYMFNTPEGRVKNP 254
Query: 85 NFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
F+K+VSS C+KED L+VGCQSG RS AT++LLA+GFK+V+NMGGGY+EWV + PV
Sbjct: 255 EFLKEVSSLCNKEDHLIVGCQSGVRSVYATADLLAEGFKDVYNMGGGYLEWVKKEFPV 428
>TC86665 similar to GP|7594903|dbj|BAA88985.2 Ntdin {Nicotiana tabacum},
partial (60%)
Length = 821
Score = 109 bits (273), Expect = 3e-25
Identities = 57/113 (50%), Positives = 69/113 (60%)
Frame = +1
Query: 32 TIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVS 91
++ V A L+ GH YLDVRT EEF GH A INIPY+ G KN NFVK+VS
Sbjct: 355 SVPVRVAHELLLAGHKYLDVRTTEEFNAGH--APGAINIPYMYKVGSGMTKNSNFVKEVS 528
Query: 92 SSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPVIQ 144
S KED ++VGCQ GKRS A ++LLA GF + ++ GGY W N LP Q
Sbjct: 529 SHFRKEDEVIVGCQLGKRSMMAATDLLAAGFTGLTDIAGGYAAWTQNGLPTEQ 687
>BE203893 similar to GP|20197696|gb| senescence-associated protein
{Arabidopsis thaliana}, partial (12%)
Length = 643
Score = 89.0 bits (219), Expect = 6e-19
Identities = 48/89 (53%), Positives = 61/89 (67%), Gaps = 1/89 (1%)
Frame = +1
Query: 30 VVTIDVHAAKNLIQT-GHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVK 88
VVT+DVHA K+L+ + G+ YLDVRTVEEF K HV+ IN+PYL T +GRVKN +FV
Sbjct: 172 VVTLDVHATKDLLDSSGYNYLDVRTVEEFNKSHVE--NAINVPYLFSTEEGRVKNPDFVN 345
Query: 89 QVSSSCDKEDCLVVGCQSGKRSFSATSEL 117
QV + ED L+V C +G RS A +L
Sbjct: 346 QVEAIYKSEDHLIVACNAGGRSSRAWVDL 432
>BG588641 similar to GP|20197696|gb| senescence-associated protein
{Arabidopsis thaliana}, partial (41%)
Length = 516
Score = 74.7 bits (182), Expect = 1e-14
Identities = 39/74 (52%), Positives = 49/74 (65%)
Frame = +2
Query: 44 TGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVSSSCDKEDCLVVG 103
+G+ YLDVRTVEEF K HV+ IN+PYL T +GRVKN +FV QV + ED L+V
Sbjct: 65 SGYNYLDVRTVEEFNKSHVE--NAINVPYLFSTEEGRVKNPDFVNQVEAIYKSEDHLIVA 238
Query: 104 CQSGKRSFSATSEL 117
C +G RS A +L
Sbjct: 239 CNAGGRSSRAWVDL 280
>TC78039 similar to PIR|T05875|T05875 hypothetical protein T29A15.190 -
Arabidopsis thaliana, partial (69%)
Length = 1080
Score = 45.1 bits (105), Expect = 1e-05
Identities = 33/134 (24%), Positives = 53/134 (38%), Gaps = 31/134 (23%)
Frame = +1
Query: 43 QTGHIYLDVRTVEEFEKGH------VDATKIINIPYLLDTPK-----------GRVKNLN 85
+ + LDVR EF++GH V ++I D + G +N
Sbjct: 472 ENNFVILDVRPEAEFKEGHPPDAINVQVYRLIKEWTAWDIARRAAFAFFGIFSGTEENPE 651
Query: 86 FVKQVSSSCDKEDCLVVGCQSG--------------KRSFSATSELLADGFKNVHNMGGG 131
F+K V DK ++V C +G RS A L+ +G+ NV ++ GG
Sbjct: 652 FIKSVGEQLDKNAKIIVACSAGGTMKPTQNLPQGQQSRSLIAAYLLVLNGYNNVFHLKGG 831
Query: 132 YMEWVSNKLPVIQQ 145
+W LP + +
Sbjct: 832 LYKWFKEDLPAVAE 873
>TC86666 similar to GP|7594903|dbj|BAA88985.2 Ntdin {Nicotiana tabacum},
partial (26%)
Length = 787
Score = 41.2 bits (95), Expect = 1e-04
Identities = 21/40 (52%), Positives = 24/40 (59%)
Frame = +2
Query: 40 NLIQTGHIYLDVRTVEEFEKGHVDATKIINIPYLLDTPKG 79
+L+ GH YLDVRT EEF GH A INIPY+ G
Sbjct: 524 SLLLAGHKYLDVRTTEEFNAGH--APGXINIPYMYKVGSG 637
>TC87052 similar to GP|15010630|gb|AAK73974.1 At2g42220/T24P15.13
{Arabidopsis thaliana}, partial (77%)
Length = 993
Score = 40.4 bits (93), Expect = 3e-04
Identities = 35/119 (29%), Positives = 53/119 (44%), Gaps = 22/119 (18%)
Frame = +1
Query: 29 KVVTIDVHAAKNLIQT-GHIYLDVRTVEEFEKGHVDATKIINI----------PYLLDTP 77
K VT D AK L++ G+ LDVR ++E+ H+ + + +LL T
Sbjct: 160 KFVTAD--DAKELVKVDGYNVLDVRDKSQYERAHIKTCYHVPLFVENTDNDPGTFLLRTV 333
Query: 78 KGRVKNL-----------NFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNV 125
L +FV+ V S E L++ CQ G RS +A ++L GF+NV
Sbjct: 334 HNNFSGLFFGIPFTRPNPDFVQSVKSQIQPETKLLIVCQEGLRSAAAANKLEDAGFQNV 510
>TC89250 weakly similar to GP|21592651|gb|AAM64600.1 rhodanese-like family
protein {Arabidopsis thaliana}, partial (78%)
Length = 1039
Score = 35.8 bits (81), Expect = 0.006
Identities = 35/134 (26%), Positives = 57/134 (42%), Gaps = 24/134 (17%)
Frame = +3
Query: 23 LCSSGAKVVTIDVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATKIINIP-YLLDTPKGRV 81
L SG + A+ + G + LDVR E EK HV + +++P ++ D G +
Sbjct: 162 LIESGTIRTILPKDASTVMNSEGFVLLDVRPNWEREKAHVKGS--LHVPMFVEDKDNGPL 335
Query: 82 K----------------------NLNFVKQVSSSCD-KEDCLVVGCQSGKRSFSATSELL 118
N F+ QV + K ++V C G RS +ATS+L
Sbjct: 336 TLLKKWVHFGYIGAWTGQYLTTFNSEFLSQVENVVPGKGTKVLVACGEGLRSMTATSKLY 515
Query: 119 ADGFKNVHNMGGGY 132
G++N+ + GG+
Sbjct: 516 NGGYRNLGWLVGGF 557
>TC81501
Length = 716
Score = 27.3 bits (59), Expect = 2.2
Identities = 25/85 (29%), Positives = 37/85 (43%), Gaps = 1/85 (1%)
Frame = +3
Query: 59 KGHVDATKIINIPYLLDTPKGRVKNLNFVK-QVSSSCDKEDCLVVGCQSGKRSFSATSEL 117
KG +DA I + Y ++ + F+K Q S S DK+ CL V + T +
Sbjct: 138 KG*LDAP*IHKLGY-------KIYVIIFLKCQ*S*S*DKDSCLKVLYKGSPNLLLKTRSV 296
Query: 118 LADGFKNVHNMGGGYMEWVSNKLPV 142
L DG+ + G Y E V + V
Sbjct: 297 LLDGYIKIQQDGDSYEEIVDELMDV 371
>BI312404
Length = 759
Score = 26.2 bits (56), Expect = 4.9
Identities = 14/48 (29%), Positives = 20/48 (41%)
Frame = +1
Query: 95 DKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
DK+ CL V + T +L DG+ + G Y E V + V
Sbjct: 337 DKDSCLKVLYKGSPNLLLKTRSVLLDGYIKIQQDGDSYEEIVDELMDV 480
>BF519432 similar to GP|6682233|gb| putative pectate lyase {Arabidopsis
thaliana}, partial (45%)
Length = 531
Score = 26.2 bits (56), Expect = 4.9
Identities = 12/33 (36%), Positives = 20/33 (60%)
Frame = +2
Query: 34 DVHAAKNLIQTGHIYLDVRTVEEFEKGHVDATK 66
DV A + + HI++D T+ +FE G +D T+
Sbjct: 365 DVDAIQIKPNSKHIWIDRCTLSDFEDGLIDITR 463
>TC83089
Length = 887
Score = 26.2 bits (56), Expect = 4.9
Identities = 19/88 (21%), Positives = 40/88 (44%), Gaps = 6/88 (6%)
Frame = +3
Query: 52 RTVEEFEKGHVDATKIINIPYLLDTPKGRVKNLNFVKQVSSSCDK------EDCLVVGCQ 105
+ + E+ +G++D+ ++ P L P+ ++++ ++C K CL+ G +
Sbjct: 258 QALREYYEGYLDSATRLSDPQNLGDPQE-------LEKIRTTCVKWYLLYLVGCLLFGDK 416
Query: 106 SGKRSFSATSELLADGFKNVHNMGGGYM 133
S KR + DG+ + N G M
Sbjct: 417 SNKRIELVYLTTMEDGYAGMRNHSWGGM 500
>TC87967 homologue to GP|12720419|gb|AAK02280.1 unknown {Pasteurella
multocida}, partial (3%)
Length = 1018
Score = 26.2 bits (56), Expect = 4.9
Identities = 14/48 (29%), Positives = 20/48 (41%)
Frame = +2
Query: 95 DKEDCLVVGCQSGKRSFSATSELLADGFKNVHNMGGGYMEWVSNKLPV 142
DK+ CL V + T +L DG+ + G Y E V + V
Sbjct: 827 DKDSCLKVLYKGSPNLLLKTRSVLLDGYIKIQQDGDSYEEIVDELMDV 970
>TC93673 similar to GP|16323198|gb|AAL15333.1 At1g15980/T24D18_8
{Arabidopsis thaliana}, partial (51%)
Length = 856
Score = 25.8 bits (55), Expect = 6.4
Identities = 12/28 (42%), Positives = 17/28 (59%)
Frame = +3
Query: 22 VLCSSGAKVVTIDVHAAKNLIQTGHIYL 49
V+ S K++ IDV A N IQT ++ L
Sbjct: 720 VISSKTGKLINIDVEAVTNAIQTFNVSL 803
>TC84729 homologue to PIR|C96615|C96615 hypothetical protein T18I24.10
[imported] - Arabidopsis thaliana, partial (0%)
Length = 661
Score = 25.8 bits (55), Expect = 6.4
Identities = 14/56 (25%), Positives = 29/56 (51%)
Frame = +1
Query: 73 LLDTPKGRVKNLNFVKQVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNVHNM 128
L TPKG N+++ K CD+ + +V K S+ + ++ + F +++N+
Sbjct: 115 LTPTPKGWSGNIHYCKWNGIRCDQSNQVVTAI---KLPSSSLTGIIPENFNSLNNL 273
>TC83479 similar to GP|23476992|emb|CAD48949. hypothetical protein
{Plasmodium falciparum 3D7}, partial (0%)
Length = 1222
Score = 25.4 bits (54), Expect = 8.4
Identities = 18/66 (27%), Positives = 31/66 (46%), Gaps = 3/66 (4%)
Frame = +2
Query: 69 NIPYLLDTPKGRVKNLNFVK---QVSSSCDKEDCLVVGCQSGKRSFSATSELLADGFKNV 125
NIPY+L + R+K + +K Q+++ + ++ C+ K T +L DG N
Sbjct: 857 NIPYVLSFTQERIKGMVLLKNLNQIANILNPVSRSIIWCEWKKPEIGWT-KLNTDGSVNK 1033
Query: 126 HNMGGG 131
G G
Sbjct: 1034ETAGFG 1051
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.137 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,852,543
Number of Sequences: 36976
Number of extensions: 64741
Number of successful extensions: 351
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 343
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 345
length of query: 145
length of database: 9,014,727
effective HSP length: 87
effective length of query: 58
effective length of database: 5,797,815
effective search space: 336273270
effective search space used: 336273270
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 53 (25.0 bits)
Medicago: description of AC147013.5


