

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147013.12 - phase: 0
(687 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
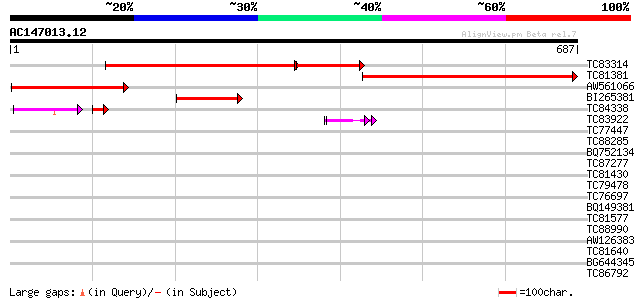
Score E
Sequences producing significant alignments: (bits) Value
TC83314 similar to GP|10177135|dbj|BAB10425. gene_id:K2A18.28~pi... 462 e-176
TC81381 similar to GP|10177135|dbj|BAB10425. gene_id:K2A18.28~pi... 386 e-107
AW561066 similar to GP|10177135|db gene_id:K2A18.28~pir||T10240~... 224 7e-59
BI265381 similar to GP|10177135|dbj gene_id:K2A18.28~pir||T10240... 76 4e-14
TC84338 weakly similar to GP|1495247|emb|CAA66220.1 orf 05 {Arab... 59 6e-10
TC83922 similar to GP|15809990|gb|AAL06922.1 AT5g60530/muf9_180 ... 45 9e-05
TC77447 similar to GP|16974501|gb|AAL31160.1 AT3g06720/F3E22_14 ... 39 0.005
TC88285 38 0.011
BQ752134 GP|7300332|gb CG7847-PA {Drosophila melanogaster}, part... 38 0.011
TC87277 similar to PIR|T45588|T45588 arm repeat containing prote... 38 0.015
TC81430 weakly similar to GP|18491179|gb|AAL69492.1 unknown prot... 37 0.019
TC79478 similar to GP|18491179|gb|AAL69492.1 unknown protein {Ar... 35 0.073
TC76697 35 0.073
BQ149381 weakly similar to GP|16209658|gb At1g61350/T1F9_16 {Ara... 35 0.096
TC81577 similar to PIR|T48012|T48012 hypothetical protein T17J13... 34 0.21
TC88990 similar to PIR|T11751|T11751 transcription repressor ROM... 33 0.28
AW126383 weakly similar to SP|P34480|NAGA_ Putative N-acetylgluc... 33 0.36
TC81640 similar to GP|15292863|gb|AAK92802.1 unknown protein {Ar... 33 0.47
BG644345 weakly similar to GP|19697333|gb putative protein poten... 33 0.47
TC86792 similar to GP|10281004|dbj|BAB13742. pseudo-response reg... 32 0.62
>TC83314 similar to GP|10177135|dbj|BAB10425.
gene_id:K2A18.28~pir||T10240~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (26%)
Length = 956
Score = 462 bits (1189), Expect(2) = e-176
Identities = 231/235 (98%), Positives = 233/235 (98%)
Frame = +3
Query: 117 WLLRVSAPADDRGGEYLGLPPIAANEPILCFIWEQIAMLFTGSQEVRSDAAASLVSLARG 176
WLLRVSAPADDRGGEYLGLPPIAANEPILCFIWEQIAMLFTGSQEVRSDAAASLVSLARG
Sbjct: 12 WLLRVSAPADDRGGEYLGLPPIAANEPILCFIWEQIAMLFTGSQEVRSDAAASLVSLARG 191
Query: 177 SDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIGLLGRDAESVEHMIHVGVCSVFAK 236
SDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIGLLGRDAESVEHMIHVGVCSVFAK
Sbjct: 192 SDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIGLLGRDAESVEHMIHVGVCSVFAK 371
Query: 237 ILKEGPMKVQGVVAWAVSELAANYPKCQELFAQHNIIRLLVGHLAFETVEEHSKYAIVSM 296
ILKEGPMKVQGVVAWAVSELAANYPKCQELFAQHNIIRLLVGHLAFETVEEHSKYAIVSM
Sbjct: 372 ILKEGPMKVQGVVAWAVSELAANYPKCQELFAQHNIIRLLVGHLAFETVEEHSKYAIVSM 551
Query: 297 KANSIHAAVVMASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPR 351
KANSIHAAVVMASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPL E+ +
Sbjct: 552 KANSIHAAVVMASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLSEKTK 716
Score = 176 bits (447), Expect(2) = e-176
Identities = 82/83 (98%), Positives = 83/83 (99%)
Frame = +1
Query: 348 ERPRNLHRVITSTMAIHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQ 407
+RPRNLHRVITSTMAIHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQ
Sbjct: 706 KRPRNLHRVITSTMAIHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQ 885
Query: 408 GNHNNHQRNYSHSGINMKGRESE 430
GNHNNHQRNYSHSGINMKGRESE
Sbjct: 886 GNHNNHQRNYSHSGINMKGRESE 954
>TC81381 similar to GP|10177135|dbj|BAB10425.
gene_id:K2A18.28~pir||T10240~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (34%)
Length = 965
Score = 386 bits (991), Expect = e-107
Identities = 200/260 (76%), Positives = 226/260 (86%)
Frame = +3
Query: 428 ESEDAETKASMKEMAARALWHLAKGNVAICRSITESRALLCFSVLLEKGPEAVQYNSAMA 487
E EDAE+KA MK MAA+AL +LAKGN AICRSITESRALLCF++LLEKGPE V+YNSA+A
Sbjct: 3 ELEDAESKADMKAMAAKALRYLAKGNSAICRSITESRALLCFAILLEKGPEEVKYNSALA 182
Query: 488 LMEITAVAEKDAELRKSAFKPNSPACKAVVDQVLKIIEKADSDLLIPCVKAIGNLARTFK 547
L EITAVAEKD ELR+SAFKPN+PACKAVVDQV+ II+K D LLIPC+K IG+LARTF+
Sbjct: 183 LKEITAVAEKDPELRRSAFKPNTPACKAVVDQVIDIIDKEDKRLLIPCIKVIGSLARTFR 362
Query: 548 ATETRMIGPLVKLLDEREAEVSREASIALRKFAGSENYLHVDHSNAIISAGGAKHLIQLV 607
ATETR+IGPLV+LLDEREAEVS+EA+ +L KFA ++NYLH+DH AIIS GG K L+QLV
Sbjct: 363 ATETRIIGPLVRLLDEREAEVSKEAADSLAKFASNDNYLHLDHCKAIISFGGVKPLVQLV 542
Query: 608 YFGEQMVQIPALVLLSYIALHVPDSEELALAEVLGVLEWASKQSFMQHDETLEELLQEAK 667
Y GE VQ ALVLLSYIALHVPDSEELA AE+LGVLEWASKQ M HDE +E LLQE+K
Sbjct: 543 YLGEPPVQYSALVLLSYIALHVPDSEELAKAEILGVLEWASKQPNMAHDEAIEALLQESK 722
Query: 668 SRLELYQSRGSRGFHHKLHQ 687
SRLELYQSRGSRGF KLHQ
Sbjct: 723 SRLELYQSRGSRGF-QKLHQ 779
>AW561066 similar to GP|10177135|db gene_id:K2A18.28~pir||T10240~strong
similarity to unknown protein {Arabidopsis thaliana},
partial (19%)
Length = 432
Score = 224 bits (572), Expect = 7e-59
Identities = 115/142 (80%), Positives = 133/142 (92%), Gaps = 1/142 (0%)
Frame = +3
Query: 3 DIVKQILAKPIQLADQVTKAADE-ASSFKQECSELKSKTEKLATLLRQAARASSDLYERP 61
DIVKQ+LAKPIQLADQV+KAA+E +SSFKQEC +LKSKTEKLA+LLRQAAR+SSDLYERP
Sbjct: 6 DIVKQLLAKPIQLADQVSKAAEEGSSSFKQECLDLKSKTEKLASLLRQAARSSSDLYERP 185
Query: 62 TKRIIEETEQVLDKALSLVLKCRANGLMKRVFTIIPAAAFRKTSSHLENSIGDVSWLLRV 121
T+RII +TEQVL+KAL+LVLKC+ NGLMKRVF+I+P+AAFRK SSHLENSIGDVSWLLRV
Sbjct: 186 TRRIIGDTEQVLEKALTLVLKCKVNGLMKRVFSIVPSAAFRKMSSHLENSIGDVSWLLRV 365
Query: 122 SAPADDRGGEYLGLPPIAANEP 143
SAPA++ E LGLPPIA+NEP
Sbjct: 366 SAPAEEGSYECLGLPPIASNEP 431
>BI265381 similar to GP|10177135|dbj gene_id:K2A18.28~pir||T10240~strong
similarity to unknown protein {Arabidopsis thaliana},
partial (10%)
Length = 248
Score = 76.3 bits (186), Expect = 4e-14
Identities = 42/81 (51%), Positives = 52/81 (63%), Gaps = 1/81 (1%)
Frame = +2
Query: 203 DGQENAARAIGLLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWA-VSELAANYP 261
+GQENAA+AIGLLGRD ESVE MIH F + + ++ VSEL + YP
Sbjct: 2 EGQENAAKAIGLLGRDPESVEVMIHAWCLFCFREDS*RRSYESSSCCSFGPVSELVSKYP 181
Query: 262 KCQELFAQHNIIRLLVGHLAF 282
KCQ++FAQHNI+RLLV F
Sbjct: 182 KCQDVFAQHNIVRLLVSXHCF 244
>TC84338 weakly similar to GP|1495247|emb|CAA66220.1 orf 05 {Arabidopsis
thaliana}, partial (9%)
Length = 670
Score = 59.3 bits (142), Expect(2) = 6e-10
Identities = 33/88 (37%), Positives = 51/88 (57%), Gaps = 4/88 (4%)
Frame = +1
Query: 5 VKQILAKPIQLADQVTKAADEASSFKQECSELKSKTEKLATLLRQAAR----ASSDLYER 60
+++ L PI + +QV K A+EA S K EC EL K E L LR R + L +R
Sbjct: 229 LEEELLIPILMGEQVIKLAEEAKSSKVECRELAKKVEILCKNLRGVVRVVTGSHQSLNDR 408
Query: 61 PTKRIIEETEQVLDKALSLVLKCRANGL 88
P +R++ E + L+K L+LV +C++ G+
Sbjct: 409 PIRRMVRELSKNLEKTLALVRRCKSQGV 492
Score = 22.7 bits (47), Expect(2) = 6e-10
Identities = 9/19 (47%), Positives = 14/19 (73%)
Frame = +2
Query: 101 FRKTSSHLENSIGDVSWLL 119
FRK + LE+S GD+ W++
Sbjct: 536 FRKVWNLLESSNGDMMWVV 592
>TC83922 similar to GP|15809990|gb|AAL06922.1 AT5g60530/muf9_180
{Arabidopsis thaliana}, partial (21%)
Length = 619
Score = 45.1 bits (105), Expect = 9e-05
Identities = 23/51 (45%), Positives = 27/51 (52%)
Frame = +1
Query: 385 NSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESEDAETK 435
N N NGNG GNGNGNGN+G +GN + KG+E E A K
Sbjct: 118 NGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKE--------KGKEKEKAPKK 246
Score = 43.5 bits (101), Expect = 3e-04
Identities = 23/63 (36%), Positives = 32/63 (50%)
Frame = +1
Query: 382 ILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESEDAETKASMKEM 441
+L + G GNGNGNGN+G +GN N + N N KG + E+ + K KE
Sbjct: 70 LLISMEAAIAQGQGNGNGNGNNGNGKGNGNGNGNNG-----NGKGNDKEEGKEKGKEKEK 234
Query: 442 AAR 444
A +
Sbjct: 235 APK 243
Score = 31.6 bits (70), Expect = 1.1
Identities = 18/52 (34%), Positives = 23/52 (43%)
Frame = +1
Query: 356 VITSTMAIHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQ 407
++ S A A + GN N N N N NGNG GN G + GK+
Sbjct: 70 LLISMEAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKE 225
>TC77447 similar to GP|16974501|gb|AAL31160.1 AT3g06720/F3E22_14
{Arabidopsis thaliana}, partial (96%)
Length = 2271
Score = 39.3 bits (90), Expect = 0.005
Identities = 28/113 (24%), Positives = 55/113 (47%), Gaps = 2/113 (1%)
Frame = +2
Query: 184 IIEEGGVGPLLK-LIKEGKADGQENAARAI-GLLGRDAESVEHMIHVGVCSVFAKILKEG 241
+I+ G V ++ L++E Q AA A+ + +E+ + +I G +F K+L
Sbjct: 440 VIQAGVVPRFIEFLMREDFPQLQFEAAWALTNIASGTSENTKVVIEAGAVPIFVKLLASP 619
Query: 242 PMKVQGVVAWAVSELAANYPKCQELFAQHNIIRLLVGHLAFETVEEHSKYAIV 294
V+ WA+ +A + P+C++L H + L+ L EH+K +++
Sbjct: 620 SDDVREQAVWALGNVAGDSPRCRDLVLGHGALLPLLAQL-----NEHAKLSML 763
>TC88285
Length = 2273
Score = 38.1 bits (87), Expect = 0.011
Identities = 20/81 (24%), Positives = 41/81 (49%), Gaps = 2/81 (2%)
Frame = +2
Query: 184 IIEEGGVGPLLKLIKEGKADGQENAARAIGLL--GRDAESVEHMIHVGVCSVFAKILKEG 241
+IE GG+ ++ ++K GK + +ENA A+ + ES ++ G+ + L G
Sbjct: 1541 LIEMGGLDAIISILKTGKMEAKENALSALFRFTDPTNIESQRDLVKRGIYPLLVDFLNTG 1720
Query: 242 PMKVQGVVAWAVSELAANYPK 262
+ + + A + +L+ + PK
Sbjct: 1721 SVTAKAIAAAFIGDLSMSTPK 1783
>BQ752134 GP|7300332|gb CG7847-PA {Drosophila melanogaster}, partial (2%)
Length = 758
Score = 38.1 bits (87), Expect = 0.011
Identities = 17/63 (26%), Positives = 28/63 (43%)
Frame = +2
Query: 364 HAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGIN 423
H + ++ A + + A+ N N N N N + + G H+NH+R +SH G
Sbjct: 416 HIPAMSYSDALAAAREEEAKASHNNNNNNNNNNNNNSSSSGSGGQEHSNHKRQHSHGGAT 595
Query: 424 MKG 426
G
Sbjct: 596 ESG 604
>TC87277 similar to PIR|T45588|T45588 arm repeat containing protein homolog
- Arabidopsis thaliana, partial (48%)
Length = 1393
Score = 37.7 bits (86), Expect = 0.015
Identities = 23/75 (30%), Positives = 36/75 (47%)
Frame = +1
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
+ + GS + +AAA LV L G +Y E G + PLL+L + G G+ A +
Sbjct: 781 VKFIGNGSPRNKENAAAVLVHLCSGDRQYIAQAQELGVMAPLLELAQNGTDRGKRKATQL 960
Query: 212 IGLLGRDAESVEHMI 226
I + R E +H +
Sbjct: 961 IERMSRIREQEQHEV 1005
>TC81430 weakly similar to GP|18491179|gb|AAL69492.1 unknown protein
{Arabidopsis thaliana}, partial (49%)
Length = 1491
Score = 37.4 bits (85), Expect = 0.019
Identities = 36/132 (27%), Positives = 55/132 (41%)
Frame = +2
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
I ML + ++R +A +L LA+ + I GG+ PLLKL+ Q NAA A
Sbjct: 80 IEMLSSPDVQLREMSAFALGRLAQDTHNQAG-IAHNGGLVPLLKLLDSKNGSLQHNAAFA 256
Query: 212 IGLLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVSELAANYPKCQELFAQHN 271
+ L + ++V + I VG + L+EG VQ L K H
Sbjct: 257 LYGLAENEDNVSYFIRVG----GVQRLQEGEFIVQATKDCVAKTLKRLEEKIHGRVLNHL 424
Query: 272 IIRLLVGHLAFE 283
+ + V F+
Sbjct: 425 LYLMRVSERGFQ 460
>TC79478 similar to GP|18491179|gb|AAL69492.1 unknown protein {Arabidopsis
thaliana}, partial (39%)
Length = 958
Score = 35.4 bits (80), Expect = 0.073
Identities = 43/161 (26%), Positives = 65/161 (39%), Gaps = 32/161 (19%)
Frame = +2
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
I ML + +++ +A +L LA+ + I GG+ PLLKL+ Q NAA A
Sbjct: 437 IEMLQSSDVQLKEMSAFALGRLAQDTHNQAG-IAHSGGLVPLLKLLDSKNGSLQHNAAFA 613
Query: 212 IGLLGRDAESVEHMIHVGVCSVF--------------AKILKEGPMKVQGVV------AW 251
+ L + ++V I +G F AK LK K+ G V
Sbjct: 614 LYGLAENEDNVPDFIRIGGIKRFQDGEFIIQATKDCVAKTLKRLEEKINGRVLNHLLYLM 793
Query: 252 AVSELA------------ANYPKCQELFAQHNIIRLLVGHL 280
VSE A + +++F HN + LL+G L
Sbjct: 794 RVSEKAFQRRVALALAHLCSADDQKKIFIDHNGLELLIGLL 916
>TC76697
Length = 922
Score = 35.4 bits (80), Expect = 0.073
Identities = 27/88 (30%), Positives = 39/88 (43%), Gaps = 11/88 (12%)
Frame = +1
Query: 364 HAASKQPNEGNEANQNQNILANSNTPNGNGLGNGN-GNGNDG----GKQGNHNNHQ---- 414
H+ SK G+ N N+ + NG+G GN N G+GN G H+ H
Sbjct: 247 HSFSKHHGHGSNIGGNHNMFHQDSMSNGHGFGNNNHGHGNGQKFPFGATNKHSPHHGGGV 426
Query: 415 RNYSH--SGINMKGRESEDAETKASMKE 440
R ++H G N E E+ E +A +E
Sbjct: 427 RPFNHHGGGHNDYVSEHEEYEFEAYKEE 510
>BQ149381 weakly similar to GP|16209658|gb At1g61350/T1F9_16 {Arabidopsis
thaliana}, partial (18%)
Length = 400
Score = 35.0 bits (79), Expect = 0.096
Identities = 20/67 (29%), Positives = 37/67 (54%), Gaps = 1/67 (1%)
Frame = +3
Query: 156 FTGSQEVR-SDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIGL 214
F GS EV + +A +V + G D Y ++I G + PL++++ G G+ AAR +
Sbjct: 54 FLGSNEVEIQEESAKVVCVLAGFDSYKGVLISAGVIAPLIRVLDCGSELGKVAAARCLMK 233
Query: 215 LGRDAES 221
L ++++
Sbjct: 234 LTENSDN 254
>TC81577 similar to PIR|T48012|T48012 hypothetical protein T17J13.160 -
Arabidopsis thaliana, partial (20%)
Length = 1491
Score = 33.9 bits (76), Expect = 0.21
Identities = 20/56 (35%), Positives = 26/56 (45%), Gaps = 7/56 (12%)
Frame = +3
Query: 371 NEGNEANQNQNI-----LANSNTPNGNGLGNGNGN--GNDGGKQGNHNNHQRNYSH 419
N N N N+ AN N + L + NGN GN +QGNH N Q N+ +
Sbjct: 480 NNYRPGNYNPNVSQSGSTANFERANSDPLWSNNGNQQGNHQNQQGNHQNQQGNHQN 647
>TC88990 similar to PIR|T11751|T11751 transcription repressor ROM1 - kidney
bean, partial (91%)
Length = 1528
Score = 33.5 bits (75), Expect = 0.28
Identities = 25/105 (23%), Positives = 46/105 (43%), Gaps = 16/105 (15%)
Frame = +1
Query: 338 HGRVSHHPLGERPRNLHR---VITSTMAIHAASKQPNEG------NEANQNQNILANSNT 388
+G +P P N++ ++ + A+H ++ +G + + + + AN++
Sbjct: 430 YGTPVPYPAMFPPGNIYAHPSMVVTPSAMHQTTEFEGKGPDGKDKDSSKKPKGTSANTSA 609
Query: 389 PNGNGLGNGNGNGNDG-------GKQGNHNNHQRNYSHSGINMKG 426
G G G+G+GNDG G +G+ N N S N KG
Sbjct: 610 KAGEGGKAGSGSGNDGFSHSGDSGSEGSSNASDENQQESARNKKG 744
>AW126383 weakly similar to SP|P34480|NAGA_ Putative
N-acetylglucosamine-6-phosphate deacetylase (EC
3.5.1.25) (GlcNAc 6-P deacetylase)., partial (11%)
Length = 525
Score = 33.1 bits (74), Expect = 0.36
Identities = 15/29 (51%), Positives = 19/29 (64%), Gaps = 1/29 (3%)
Frame = +1
Query: 375 EANQNQNILANSN-TPNGNGLGNGNGNGN 402
+ + +I+ N N NGNG GNGNGNGN
Sbjct: 97 DITKKTHIIQNGNMNGNGNGNGNGNGNGN 183
>TC81640 similar to GP|15292863|gb|AAK92802.1 unknown protein {Arabidopsis
thaliana}, partial (40%)
Length = 810
Score = 32.7 bits (73), Expect = 0.47
Identities = 24/57 (42%), Positives = 33/57 (57%)
Frame = +2
Query: 166 AAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAIGLLGRDAESV 222
A L SLA G D + I+EEGG+ L++ I++G G+E A + LL AESV
Sbjct: 356 AMVVLNSLA-GFDEGKEAIVEEGGIAALVEAIEDGSVKGKEFA--VLTLLQLCAESV 517
>BG644345 weakly similar to GP|19697333|gb putative protein potential
transcriptional repressor Not4hp - Mus musculus, partial
(7%)
Length = 635
Score = 32.7 bits (73), Expect = 0.47
Identities = 19/51 (37%), Positives = 23/51 (44%), Gaps = 3/51 (5%)
Frame = +2
Query: 376 ANQNQNILANSNTPNGNGLGNGN---GNGNDGGKQGNHNNHQRNYSHSGIN 423
+N N N +NSN N N N N N N+ N+NN N SH N
Sbjct: 320 SNNNFNFSSNSNNNNNNFSNNNNNNFSNNNNCNCSNNNNNCCSNISHHSSN 472
>TC86792 similar to GP|10281004|dbj|BAB13742. pseudo-response regulator 7
{Arabidopsis thaliana}, partial (32%)
Length = 2209
Score = 32.3 bits (72), Expect = 0.62
Identities = 18/75 (24%), Positives = 33/75 (44%), Gaps = 4/75 (5%)
Frame = +1
Query: 358 TSTMAIHAASKQPNEGNEANQNQNILANS---NTPNGNGLGNGNGNGN-DGGKQGNHNNH 413
T +I+ ++ N G+ + N+ N N NGL +G+G+ G GN +
Sbjct: 1591 TGNYSINRSASGSNHGSNGQNGSSTAINAGGTNIDNNNGLAGNSGSGDASGNGSGNRVDQ 1770
Query: 414 QRNYSHSGINMKGRE 428
+N+ +K R+
Sbjct: 1771 SKNFQREAALIKFRQ 1815
Score = 29.3 bits (64), Expect = 5.3
Identities = 18/54 (33%), Positives = 24/54 (44%)
Frame = +1
Query: 368 KQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSG 421
K P EGN N + N A+ + NG + N GG ++NN S SG
Sbjct: 1570 KGPVEGNTGNYSINRSASGSNHGSNGQNGSSTAINAGGTNIDNNNGLAGNSGSG 1731
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.131 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,307,395
Number of Sequences: 36976
Number of extensions: 230958
Number of successful extensions: 1362
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 1203
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1297
length of query: 687
length of database: 9,014,727
effective HSP length: 103
effective length of query: 584
effective length of database: 5,206,199
effective search space: 3040420216
effective search space used: 3040420216
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147013.12


