

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147012.5 + phase: 0
(789 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
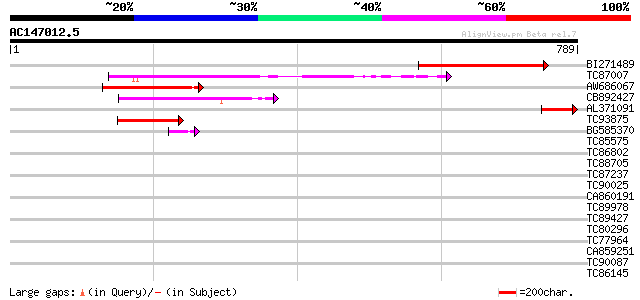
Score E
Sequences producing significant alignments: (bits) Value
BI271489 weakly similar to PIR|E86457|E8 probable RNA helicase ... 366 e-101
TC87007 homologue to PIR|T48023|T48023 ATP-dependent RNA helicas... 154 1e-37
AW686067 similar to GP|22535591|d putative DEIH-box RNA/DNA heli... 114 1e-25
CB892427 similar to PIR|G96613|G9 hypothetical protein T15M6.7 [... 103 3e-22
AL371091 97 3e-20
TC93875 similar to PIR|T49915|T49915 pre-mRNA splicing factor AT... 96 7e-20
BG585370 similar to GP|10440614|gb| putative ATP-dependent RNA h... 46 6e-05
TC85575 similar to PIR|D86410|D86410 protein F3M18.16 [imported]... 42 0.001
TC86802 similar to GP|7939577|dbj|BAA95778.1 ATP-dependent RNA h... 42 0.001
TC88705 similar to GP|10177834|dbj|BAB11263. gene_id:MCD7.9~unkn... 40 0.003
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 39 0.008
TC90025 similar to GP|18855061|gb|AAL79753.1 putative RNA helica... 36 0.065
CA860191 homologue to GP|19697342|gb hypothetical protein {Dicty... 36 0.065
TC89978 similar to GP|12324575|gb|AAG52236.1 putative DnaJ prote... 35 0.11
TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed pr... 34 0.19
TC80296 similar to PIR|H86265|H86265 protein F3F19.18 [imported]... 34 0.25
TC77964 homologue to GP|14190381|gb|AAK55671.1 AT5g11200/F2I11_9... 33 0.32
CA859251 similar to GP|21305823|gb DNA polymerase I {Hz-1 insect... 33 0.32
TC90087 similar to PIR|T05841|T05841 spliceosome-associated prot... 33 0.42
TC86145 homologue to GP|10334499|emb|CAC10211. hypothetical prot... 33 0.42
>BI271489 weakly similar to PIR|E86457|E8 probable RNA helicase 27866-23496
[imported] - Arabidopsis thaliana, partial (12%)
Length = 544
Score = 366 bits (940), Expect = e-101
Identities = 175/181 (96%), Positives = 177/181 (97%)
Frame = +1
Query: 570 VHPEFLVYNELLETKRPNKEGETSAKRAYMHGVTNVDPTWLVENAKSSCIFSPPLTDPRP 629
V PEFLVYNELLETKRPNKEGETSAKRAYMHGVTNVDPTWLVENAKSSCIFSPPLTDPRP
Sbjct: 1 VRPEFLVYNELLETKRPNKEGETSAKRAYMHGVTNVDPTWLVENAKSSCIFSPPLTDPRP 180
Query: 630 FYDAQADQVKCWVIPTFGRFCWELPKHSIPISNVEHRVQVFAYALLEGQVCTCLKSVRKY 689
FYDAQADQVKCWVIPTFGRFCWELPKHSIPISNVEHRVQVFAYALLEGQVC CLK+VRKY
Sbjct: 181 FYDAQADQVKCWVIPTFGRFCWELPKHSIPISNVEHRVQVFAYALLEGQVCPCLKTVRKY 360
Query: 690 MSAPPETILRREALGQKRVGNLISKLNSRLIDSSAMLRIVWKQNPRELFSEILDWFQQGF 749
MSAPPETILRRE+ GQKRVGNLISKLNSRLIDSSA LRIVWKQNPRELFSEILDWFQQGF
Sbjct: 361 MSAPPETILRRESFGQKRVGNLISKLNSRLIDSSATLRIVWKQNPRELFSEILDWFQQGF 540
Query: 750 R 750
R
Sbjct: 541 R 543
>TC87007 homologue to PIR|T48023|T48023 ATP-dependent RNA helicase-like
protein - Arabidopsis thaliana, partial (96%)
Length = 2361
Score = 154 bits (390), Expect = 1e-37
Identities = 134/486 (27%), Positives = 223/486 (45%), Gaps = 9/486 (1%)
Frame = +3
Query: 138 KIARE--NHDSSPGALFVLPLYAMLPAAAQLRVFDG----VKEGE---RLVVVATNVAET 188
KI++E N G + +PLY+ LP A Q ++F+ VKEG R +VV+TN+AET
Sbjct: 1041 KISKEVANMGDQVGPVKAVPLYSTLPPAMQQKIFEPAPPPVKEGGPPGRKIVVSTNIAET 1220
Query: 189 SLTIPGIKYVVDTGREKVKNYDSSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYS 248
SLTI GI YV+D G K K Y+ +E+ V ISKASA QR+GRAGRT G C+RLY+
Sbjct: 1221 SLTIDGIVYVIDPGFAKQKVYNPRVRVESLLVSTISKASAHQRSGRAGRTQPGKCFRLYT 1400
Query: 249 SAAFSNEFPEFSPAEVEKVPVHGVVLLLKSMQIKKVANFPFPTSLKAASLLEAENCLRAL 308
+F+N+ + E+ + + VL LK + I + +F F +L+ A L L
Sbjct: 1401 ERSFNNDLQPQTYPEILRSNLANTVLTLKKLGIDDLVHFDFMDPPAPETLMRALEVLNYL 1580
Query: 309 EALDSKDELTLLGKAMALYPLSPRHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAALSL 368
ALD + LT LG+ M+ +PL P+ S+M++ + I ++ +A LS+
Sbjct: 1581 GALDDEGNLTKLGEIMSEFPLDPQMSKMLVVSPEFNCSNEI----------LSISAMLSV 1730
Query: 369 PNPFVMQYEGNDSNKDSETSEKSRMGDNENNIDKTEKTKRKKLKQTSKVAREKFRIVSSD 428
PN F+ + ++ ++ + A+ +F + D
Sbjct: 1731 PNCFI---------------------------------RPREAQKAADEAKARFGHIDGD 1811
Query: 429 ALAIAYALQCFEHSQNSVQFCEDNALHFKTMDEMSKLRQQLLRLVFFQNDKGGLEQEYSW 488
L + ++ + +C DN ++ + + +RQQL+R++ N K
Sbjct: 1812 HLTLLNVYHAYKQNNEDASWCYDNFVNNRALKSADNVRQQLVRIMARFNLK--------- 1964
Query: 489 THVTLEDVEHVWRVSSAHYPLPLVEERLICRAICAGWADRVAKRIPISKAVDGETISRAG 548
+ D +S Y + I +A+ AG+ +VA + R G
Sbjct: 1965 --LCSTD------FNSRDYYVN------IRKAMLAGYFMQVAH------------LERTG 2066
Query: 549 RYQSCMVDESIFIHRWSSVSTVHPEFLVYNELLETKRPNKEGETSAKRAYMHGVTNVDPT 608
Y + ++ + +H S+ PE+++YNE + T R ++ VT++
Sbjct: 2067 HYLTVKDNQVVHLHP-SNCLDHKPEWVIYNEYVLT-----------SRNFIRTVTDIRGE 2210
Query: 609 WLVENA 614
WLV+ A
Sbjct: 2211 WLVDIA 2228
>AW686067 similar to GP|22535591|d putative DEIH-box RNA/DNA helicase {Oryza
sativa (japonica cultivar-group)}, partial (15%)
Length = 663
Score = 114 bits (286), Expect = 1e-25
Identities = 59/140 (42%), Positives = 86/140 (61%)
Frame = +2
Query: 130 DQSKVGREKIARENHDSSPGALFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETS 189
D REK+ + ++P V+ L++M+P Q +VF G R +V++TN+AET+
Sbjct: 134 DDINRAREKLLASSFFNNPSKFVVISLHSMVPTLEQKKVFKRPPPGCRKIVLSTNLAETA 313
Query: 190 LTIPGIKYVVDTGREKVKNYDSSNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSS 249
+TI I YV+DTGR K K+YD N + T + WISKASA QR GRAGR G CY LYS
Sbjct: 314 VTIDDIVYVIDTGRMKEKSYDPYNNVSTLQSSWISKASAKQREGRAGRCQPGICYHLYSK 493
Query: 250 AAFSNEFPEFSPAEVEKVPV 269
++ P+F E++++P+
Sbjct: 494 LRAAS-LPDFQTPELKRMPI 550
>CB892427 similar to PIR|G96613|G9 hypothetical protein T15M6.7 [imported] -
Arabidopsis thaliana, partial (15%)
Length = 809
Score = 103 bits (256), Expect = 3e-22
Identities = 72/225 (32%), Positives = 118/225 (52%), Gaps = 3/225 (1%)
Frame = +2
Query: 152 FVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYDS 211
+V+PL++ + + Q +VF R VV+ATN+AETS+TI + YV+D G+ K Y+
Sbjct: 5 WVIPLHSSVASTEQKKVFLRPPGNIRKVVIATNIAETSITIDDVIYVIDCGKHKENRYNP 184
Query: 212 SNGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNEFPEFSPAEVEKVPVHG 271
+ + WIS+A+A QR GRAGR G C+RLY+ F + E+ ++P+
Sbjct: 185 QKKLSSMVEDWISQANARQRQGRAGRVKPGICFRLYTRYRFEKLMRPYQVPEMLRMPLVE 364
Query: 272 VVLLLKSMQIKKVANFPFPTSL---KAASLLEAENCLRALEALDSKDELTLLGKAMALYP 328
+ L +K + + + F T+L K ++ A + L + AL+ +ELT LG +A P
Sbjct: 365 LCLQIKLLSLGYIKPF-LSTALEPPKIEAIDSAMSLLYEVGALEGDEELTPLGHHLAKLP 541
Query: 329 LSPRHSRMILTVIKNTRYKHIRNSSLLLAYAVAAAAALSLPNPFV 373
+ +M+L Y I L+ ++ +A LS +PFV
Sbjct: 542 VDVLIGKMML-------YGAIFG---CLSPILSVSAFLSYKSPFV 646
>AL371091
Length = 500
Score = 96.7 bits (239), Expect = 3e-20
Identities = 45/50 (90%), Positives = 48/50 (96%)
Frame = +3
Query: 740 EILDWFQQGFRKHFEELWLQMLGEVLQETQEQPLHKSLKRKSKVKSKLRR 789
EILDWFQQGFRKHFEELWLQMLGEVLQETQE+PLHKS K+KSKVKSK R+
Sbjct: 3 EILDWFQQGFRKHFEELWLQMLGEVLQETQERPLHKSSKKKSKVKSKSRQ 152
>TC93875 similar to PIR|T49915|T49915 pre-mRNA splicing factor ATP-dependent
RNA helicase-like protein - Arabidopsis thaliana, partial
(33%)
Length = 1239
Score = 95.5 bits (236), Expect = 7e-20
Identities = 46/91 (50%), Positives = 65/91 (70%)
Frame = +1
Query: 151 LFVLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYD 210
L +LP+Y+ LPA Q ++F ++G R +VATN+AETSLT+ GI +V+DTG K+K Y+
Sbjct: 967 LLILPIYSQLPADLQAKIFQKAEDGARKCIVATNIAETSLTVDGIFFVIDTGYGKMKVYN 1146
Query: 211 SSNGMETYEVKWISKASAAQRAGRAGRTAAG 241
GM+ +V +S+A+A QRAGRAGRT G
Sbjct: 1147 PRMGMDALQVFPVSRAAADQRAGRAGRTGPG 1239
>BG585370 similar to GP|10440614|gb| putative ATP-dependent RNA helicase
{Oryza sativa}, partial (6%)
Length = 165
Score = 45.8 bits (107), Expect = 6e-05
Identities = 23/43 (53%), Positives = 26/43 (59%)
Frame = +2
Query: 222 WISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNEFPEFSPAEV 264
WISKASA QR GRAGR G CYRLY + PE+ E+
Sbjct: 35 WISKASARQRRGRAGRVQPGVCYRLYPK-LIHDAMPEYQLPEI 160
>TC85575 similar to PIR|D86410|D86410 protein F3M18.16 [imported] -
Arabidopsis thaliana, partial (15%)
Length = 1225
Score = 42.0 bits (97), Expect = 0.001
Identities = 54/230 (23%), Positives = 89/230 (38%), Gaps = 21/230 (9%)
Frame = +3
Query: 17 ETSSVEGININEINEAFEMPGSSSMQQTDRFS-------GYDEDDNNFDENESDSYDSET 69
+TSS + IN N++N F S + + S Y NN +N S S+
Sbjct: 342 KTSSHDEINNNKVNNYFNYQQDSDNKYPNEVSDTKLTPTSYSSSSNNNYDNNKYSSSSDD 521
Query: 70 ESELEFNDDDKNNHEGSKNNNNIVDVL---GNEGSLASLKAAFENLSGQATLSSS----- 121
S +F ++ N++E NNNN N+ S ++ K E + +++
Sbjct: 522 FSNKKFPEEGYNSNENQNNNNNDNKYFYNNNNKDSSSNTKFTEEGYNSMENRNNNNNKYF 701
Query: 122 -NVNTEDSLDQSKVGREKI-ARENHDSSPGALFVLPLYAMLPAAAQLRVFDGVKEGERLV 179
N N +DS +K E + +N +++ + + + + GER
Sbjct: 702 YNNNNKDSSSNTKFPEEGYNSMQNQNNNNYEKYNYNNKVAVNDKYSFKSNNNYNNGERQG 881
Query: 180 VVATNVAETSLTIPGIKYVVDTGREK----VKNYDSSNGMETYEVKWISK 225
+ T V E G KY D E+ N DSS G+ E W +K
Sbjct: 882 MSDTRVME------GGKYFYDVNSEEKYNPTFNGDSSKGVVNSE-NWYNK 1010
>TC86802 similar to GP|7939577|dbj|BAA95778.1 ATP-dependent RNA helicase
{Arabidopsis thaliana}, partial (52%)
Length = 2777
Score = 41.6 bits (96), Expect = 0.001
Identities = 33/121 (27%), Positives = 53/121 (43%), Gaps = 4/121 (3%)
Frame = +1
Query: 153 VLPLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYDSS 212
V L+A + A+L+ D +E + ++VAT+VA L IPG++ VV +Y
Sbjct: 1537 VWTLHAQMQQRARLKAMDRFRENDNGILVATDVAARGLDIPGVRTVV--------HYQLP 1692
Query: 213 NGMETYEVKWISKASAAQRAGRAGRTAAGHCYRLYSSAAFSNEF----PEFSPAEVEKVP 268
+ E Y R+GR R +A C S +++F FS ++ P
Sbjct: 1693 HSAEVY----------VHRSGRTARASAEGCSIALISPKETSKFASLCKSFSKDNFQRFP 1842
Query: 269 V 269
V
Sbjct: 1843 V 1845
>TC88705 similar to GP|10177834|dbj|BAB11263. gene_id:MCD7.9~unknown protein
{Arabidopsis thaliana}, partial (32%)
Length = 1685
Score = 40.0 bits (92), Expect = 0.003
Identities = 39/143 (27%), Positives = 64/143 (44%), Gaps = 22/143 (15%)
Frame = +1
Query: 5 KGSVENDSNVVNETSS--VEGININEINEAFEMPGSSSMQQTDRFSG--------YDED- 53
K SVE ++ + NE + G N NE NE + + D +G YDED
Sbjct: 541 KKSVEANTALDNEDQEDILHGTN-NEENEGYASETDDDTSKYDDDTGKYDDDTGKYDEDI 717
Query: 54 -DNNFDENE----SDSYDSETESELEFNDDDKNNHEGSKNNNNIVDVLG------NEGSL 102
D F E+E S SY S+ ESE + +DD + K+ NI+ V+ N+
Sbjct: 718 NDEEFQEDEHEDLSSSYKSDVESEPDLSDDPSWLEKIQKSVWNIIQVVNIFQTPVNQSDA 897
Query: 103 ASLKAAFENLSGQATLSSSNVNT 125
A ++ ++ S + + S +++
Sbjct: 898 ARIRKEYDESSAKLSKIQSRISS 966
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 38.9 bits (89), Expect = 0.008
Identities = 34/146 (23%), Positives = 68/146 (46%), Gaps = 4/146 (2%)
Frame = +1
Query: 7 SVENDSNVVNETSSVEGININEINEAFEMPGSSSMQQTDRFSGYDEDDNNFDENESDSYD 66
S +ND+ ++++ + N+ ++ ++ SS + G +NN DEN++DS +
Sbjct: 1297 SSQNDNANSGQSNTTSDESANDNKDSSQVTTSSE----NSAEGNSNTENNSDENQNDSKN 1464
Query: 67 SETESELEFNDDDKNNHEGSKNNNNIVDVLGNEGSL--ASLKAAFEN--LSGQATLSSSN 122
+E ++ +D N +E N NEG S+++ EN + + ++SN
Sbjct: 1465 NENTNDSGNTSNDANVNENQNENAAQTKTSENEGDAQNESVESKKENNESAHKDVDNNSN 1644
Query: 123 VNTEDSLDQSKVGREKIARENHDSSP 148
N + S D S +K +R + + P
Sbjct: 1645 SNDQGSSDTSVTQDDKESRVDLGTLP 1722
>TC90025 similar to GP|18855061|gb|AAL79753.1 putative RNA helicase {Oryza
sativa}, partial (27%)
Length = 840
Score = 35.8 bits (81), Expect = 0.065
Identities = 21/82 (25%), Positives = 41/82 (49%)
Frame = +3
Query: 155 PLYAMLPAAAQLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYDSSNG 214
PL+ + + + +G ++G V+VAT+VA L IP + ++ +Y+ +G
Sbjct: 324 PLHGDISQYQREKTLNGFRQGRFTVLVATDVASRGLDIPNVDLII--------HYELPDG 479
Query: 215 METYEVKWISKASAAQRAGRAG 236
ET+ + ++ RAG+ G
Sbjct: 480 PETF----VHRSGRTGRAGKEG 533
>CA860191 homologue to GP|19697342|gb hypothetical protein {Dictyostelium
discoideum}, partial (1%)
Length = 504
Score = 35.8 bits (81), Expect = 0.065
Identities = 15/43 (34%), Positives = 23/43 (52%)
Frame = +2
Query: 49 GYDEDDNNFDENESDSYDSETESELEFNDDDKNNHEGSKNNNN 91
G D+ N +E E + + E E FN ++ NN+ + NNNN
Sbjct: 224 GLDQTLQNDEEEEEEDEEEEDEFTTTFNSNNNNNNSNNNNNNN 352
>TC89978 similar to GP|12324575|gb|AAG52236.1 putative DnaJ protein;
34157-30943 {Arabidopsis thaliana}, partial (15%)
Length = 668
Score = 35.0 bits (79), Expect = 0.11
Identities = 14/43 (32%), Positives = 24/43 (55%)
Frame = +2
Query: 49 GYDEDDNNFDENESDSYDSETESELEFNDDDKNNHEGSKNNNN 91
G +ED++N DE D Y+SE + E + + KN ++ + N
Sbjct: 260 GVEEDEDNEDEEYDDDYESEYSEDEEDDQNSKNKNQAANGTTN 388
>TC89427 weakly similar to GP|4557063|gb|AAD22502.1| expressed protein
{Arabidopsis thaliana}, partial (50%)
Length = 753
Score = 34.3 bits (77), Expect = 0.19
Identities = 15/39 (38%), Positives = 23/39 (58%)
Frame = +1
Query: 49 GYDEDDNNFDENESDSYDSETESELEFNDDDKNNHEGSK 87
G D+DD + D+++ D+ D E E E E DDD++ K
Sbjct: 442 GSDDDDEDDDDDDDDNDDGEDEDEDEEEDDDEDQPPSKK 558
>TC80296 similar to PIR|H86265|H86265 protein F3F19.18 [imported] -
Arabidopsis thaliana, partial (34%)
Length = 1734
Score = 33.9 bits (76), Expect = 0.25
Identities = 17/64 (26%), Positives = 31/64 (47%)
Frame = +3
Query: 37 GSSSMQQTDRFSGYDEDDNNFDENESDSYDSETESELEFNDDDKNNHEGSKNNNNIVDVL 96
GS + Q+ D+ S +DDN + + S D E E +DD+ + + + + D +
Sbjct: 798 GSDNAQEDDQVSLNSDDDNQLGSDNTGSDDDEAEDHDGVSDDENDRSSDYETSGDDADNV 977
Query: 97 GNEG 100
+EG
Sbjct: 978 EDEG 989
>TC77964 homologue to GP|14190381|gb|AAK55671.1 AT5g11200/F2I11_90
{Arabidopsis thaliana}, partial (97%)
Length = 1454
Score = 33.5 bits (75), Expect = 0.32
Identities = 21/73 (28%), Positives = 35/73 (47%)
Frame = +1
Query: 165 QLRVFDGVKEGERLVVVATNVAETSLTIPGIKYVVDTGREKVKNYDSSNGMETYEVKWIS 224
+L+ + G KEG++ ++VAT++ + I + V+ NYD + +TY
Sbjct: 1060 RLKRYKGFKEGKQRILVATDLVGRGIDIERVNIVI--------NYDMPDSADTY------ 1197
Query: 225 KASAAQRAGRAGR 237
R GRAGR
Sbjct: 1198 ----LHRVGRAGR 1224
>CA859251 similar to GP|21305823|gb DNA polymerase I {Hz-1 insect virus}
[Heliothis zea virus 1], partial (2%)
Length = 803
Score = 33.5 bits (75), Expect = 0.32
Identities = 13/33 (39%), Positives = 23/33 (69%)
Frame = -1
Query: 51 DEDDNNFDENESDSYDSETESELEFNDDDKNNH 83
DE++ + DE+E+++ D E E L +N+ D N+H
Sbjct: 635 DEEEEDEDEDENENEDEEEEDWLNYNNLDSNSH 537
>TC90087 similar to PIR|T05841|T05841 spliceosome-associated protein homolog
F17L22.120 - Arabidopsis thaliana, partial (12%)
Length = 733
Score = 33.1 bits (74), Expect = 0.42
Identities = 23/106 (21%), Positives = 43/106 (39%)
Frame = +3
Query: 684 KSVRKYMSAPPETILRREALGQKRVGNLISKLNSRLIDSSAMLRIVWKQNPRELFSEILD 743
K +R PP+TIL ++ + L+SKL + W +N R+ ++
Sbjct: 228 KPLRSKTQTPPKTILMQKRILNNNNNRLLSKLKLSMYRKKQSYTKAWMKNLRKSLRNLV- 404
Query: 744 WFQQGFRKHFEELWLQMLGEVLQETQEQPLHKSLKRKSKVKSKLRR 789
R + + ++ + + Q + LKRK + SK R+
Sbjct: 405 ----SARLLIQRIMIRRMSRQKMQLQRKRQTLILKRKKMITSKKRK 530
>TC86145 homologue to GP|10334499|emb|CAC10211. hypothetical protein {Cicer
arietinum}, partial (91%)
Length = 1311
Score = 33.1 bits (74), Expect = 0.42
Identities = 22/88 (25%), Positives = 42/88 (47%), Gaps = 5/88 (5%)
Frame = +1
Query: 51 DEDDNNFDENESDSYDSETESELEFNDDDKNNHEGSKNNNNIVD-----VLGNEGSLASL 105
DEDD+ D+NE D Y+ E E + +D + G + NN +++ +E +L
Sbjct: 487 DEDDDEDDDNEED-YEDEDEDAFDVHD---HASVGDRENNPVIEFDPELFSSDEAYARAL 654
Query: 106 KAAFENLSGQATLSSSNVNTEDSLDQSK 133
+ A E L+ + ++ +D+ D +
Sbjct: 655 QEAEEREMAARLLALAGIHDQDAEDMEE 738
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.130 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,303,078
Number of Sequences: 36976
Number of extensions: 328357
Number of successful extensions: 2056
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 1904
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1988
length of query: 789
length of database: 9,014,727
effective HSP length: 104
effective length of query: 685
effective length of database: 5,169,223
effective search space: 3540917755
effective search space used: 3540917755
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147012.5


