

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147012.14 + phase: 0
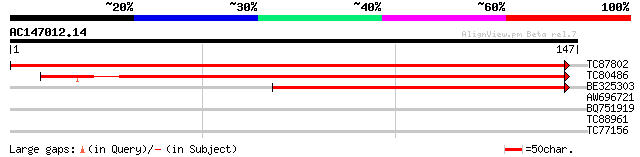
(147 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87802 similar to GP|11994617|dbj|BAB02754. contains similarity... 291 8e-80
TC80486 similar to GP|11994617|dbj|BAB02754. contains similarity... 209 4e-55
BE325303 similar to GP|11994617|dbj contains similarity to kines... 137 2e-33
AW696721 similar to GP|13536983|dbj BY-2 kinesin-like protein 10... 28 1.0
BQ751919 weakly similar to PIR|T38631|T386 aminomethyltransferas... 27 3.0
TC88961 similar to SP|Q9THX6|TL29_LYCES Putative L-ascorbate per... 27 3.9
TC77156 homologue to GP|13877717|gb|AAK43936.1 OsNAC6 protein-li... 25 8.7
>TC87802 similar to GP|11994617|dbj|BAB02754. contains similarity to kinesin
protein~gene_id:MGL6.9 {Arabidopsis thaliana}, partial
(17%)
Length = 846
Score = 291 bits (744), Expect = 8e-80
Identities = 144/145 (99%), Positives = 145/145 (99%)
Frame = +2
Query: 1 MGGQSNAAAAAALYDHAGGAVPLHPAPAGTAPDAGDAVMARWLQSAGLQHLASPLANTAI 60
MGGQSNAAAAAALYDHAGGAVPLHPAPAGTAPDAGDAVMARWLQSAGLQHLASPLANTAI
Sbjct: 389 MGGQSNAAAAAALYDHAGGAVPLHPAPAGTAPDAGDAVMARWLQSAGLQHLASPLANTAI 568
Query: 61 DQRLLPNLLMQGYGAQSAEEKQRLFKLMRSLNFNGESGSELYTPTSQTLGGAAVSDGFYS 120
DQRLLPNLLMQGYGAQSAEEKQRLFKLMRSLNFNGESGS+LYTPTSQTLGGAAVSDGFYS
Sbjct: 569 DQRLLPNLLMQGYGAQSAEEKQRLFKLMRSLNFNGESGSKLYTPTSQTLGGAAVSDGFYS 748
Query: 121 PDFRGDFGAGLLDLHAMDDTELLPE 145
PDFRGDFGAGLLDLHAMDDTELLPE
Sbjct: 749 PDFRGDFGAGLLDLHAMDDTELLPE 823
>TC80486 similar to GP|11994617|dbj|BAB02754. contains similarity to kinesin
protein~gene_id:MGL6.9 {Arabidopsis thaliana}, partial
(15%)
Length = 741
Score = 209 bits (531), Expect = 4e-55
Identities = 106/138 (76%), Positives = 117/138 (83%), Gaps = 1/138 (0%)
Frame = +1
Query: 9 AAAALYDH-AGGAVPLHPAPAGTAPDAGDAVMARWLQSAGLQHLASPLANTAIDQRLLPN 67
AAAA+YD AGG++ + DAGDAVMARWLQSAGLQHL SPLA++ +D RL PN
Sbjct: 226 AAAAVYDQPAGGSLQ------SASTDAGDAVMARWLQSAGLQHLGSPLASSGVDHRLFPN 387
Query: 68 LLMQGYGAQSAEEKQRLFKLMRSLNFNGESGSELYTPTSQTLGGAAVSDGFYSPDFRGDF 127
LLMQGYGAQSAEEKQRLFKLMR+LNFN ESGSELYTP +QTLGG + SDGFYSP+FRGDF
Sbjct: 388 LLMQGYGAQSAEEKQRLFKLMRNLNFNAESGSELYTPNTQTLGGGSASDGFYSPEFRGDF 567
Query: 128 GAGLLDLHAMDDTELLPE 145
GAGLLDLHAMDDTELL E
Sbjct: 568 GAGLLDLHAMDDTELLSE 621
>BE325303 similar to GP|11994617|dbj contains similarity to kinesin
protein~gene_id:MGL6.9 {Arabidopsis thaliana}, partial
(9%)
Length = 595
Score = 137 bits (345), Expect = 2e-33
Identities = 65/77 (84%), Positives = 71/77 (91%)
Frame = +1
Query: 69 LMQGYGAQSAEEKQRLFKLMRSLNFNGESGSELYTPTSQTLGGAAVSDGFYSPDFRGDFG 128
++ GYGAQSAEEKQRLFKLMR+LNFN ESGSELYTP +QTLGG + SDGFYSP+FRGDFG
Sbjct: 124 VLLGYGAQSAEEKQRLFKLMRNLNFNAESGSELYTPNTQTLGGGSASDGFYSPEFRGDFG 303
Query: 129 AGLLDLHAMDDTELLPE 145
AGLLDLHAMDDTELL E
Sbjct: 304 AGLLDLHAMDDTELLSE 354
>AW696721 similar to GP|13536983|dbj BY-2 kinesin-like protein 10 {Nicotiana
tabacum}, partial (11%)
Length = 687
Score = 28.5 bits (62), Expect = 1.0
Identities = 14/19 (73%), Positives = 16/19 (83%)
Frame = +1
Query: 127 FGAGLLDLHAMDDTELLPE 145
F +GLLDLH+ DTELLPE
Sbjct: 424 FTSGLLDLHSF-DTELLPE 477
>BQ751919 weakly similar to PIR|T38631|T386 aminomethyltransferase precursor
- fission yeast (Schizosaccharomyces pombe), partial
(21%)
Length = 728
Score = 26.9 bits (58), Expect = 3.0
Identities = 14/30 (46%), Positives = 14/30 (46%)
Frame = +2
Query: 12 ALYDHAGGAVPLHPAPAGTAPDAGDAVMAR 41
AL GA PLH G P GDAV R
Sbjct: 167 ALRRQPHGAAPLHRPAGGVVPRKGDAVERR 256
>TC88961 similar to SP|Q9THX6|TL29_LYCES Putative L-ascorbate peroxidase
chloroplast precursor (EC 1.11.1.11), partial (68%)
Length = 1100
Score = 26.6 bits (57), Expect = 3.9
Identities = 14/31 (45%), Positives = 17/31 (54%)
Frame = +2
Query: 73 YGAQSAEEKQRLFKLMRSLNFNGESGSELYT 103
Y AQSA + Q L +R N E G+ LYT
Sbjct: 647 YAAQSATKAQFLASAIRKCGGNEEKGNLLYT 739
>TC77156 homologue to GP|13877717|gb|AAK43936.1 OsNAC6 protein-like protein
{Arabidopsis thaliana}, partial (66%)
Length = 1096
Score = 25.4 bits (54), Expect = 8.7
Identities = 14/46 (30%), Positives = 20/46 (43%), Gaps = 7/46 (15%)
Frame = +1
Query: 16 HAGGAVPLHPAPAGTAP-------DAGDAVMARWLQSAGLQHLASP 54
H GG +P HP P TA D D++ S+ + + SP
Sbjct: 802 HVGGGLPPHPPPQTTAGIRDYMYFDTSDSIPKLHTDSSCSEQVVSP 939
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,478,673
Number of Sequences: 36976
Number of extensions: 36635
Number of successful extensions: 124
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 124
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 124
length of query: 147
length of database: 9,014,727
effective HSP length: 87
effective length of query: 60
effective length of database: 5,797,815
effective search space: 347868900
effective search space used: 347868900
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 54 (25.4 bits)
Medicago: description of AC147012.14


