

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147011.8 - phase: 0
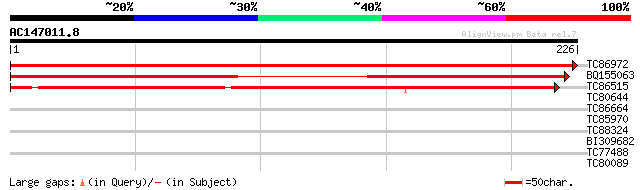
(226 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86972 similar to GP|9954743|gb|AAG09094.1| Unknown protein {Ar... 452 e-128
BQ155063 weakly similar to GP|12039349|gb putative pyrrolidone c... 317 2e-87
TC86515 similar to GP|22136908|gb|AAM91798.1 unknown protein {Ar... 288 1e-78
TC80644 similar to GP|20259627|gb|AAM14170.1 unknown protein {Ar... 28 2.2
TC86664 similar to GP|20340237|gb|AAM19705.1 protein phosphatase... 28 2.9
TC85970 homologue to PIR|T50532|T50532 ribosomal protein S27 - A... 28 3.7
TC88324 similar to PIR|F86238|F86238 protein T10O24.11 [imported... 28 3.7
BI309682 weakly similar to GP|14532612|g unknown protein {Arabid... 27 6.4
TC77488 similar to GP|15912313|gb|AAL08290.1 AT5g24690/MXC17_8 {... 27 6.4
TC80089 27 8.3
>TC86972 similar to GP|9954743|gb|AAG09094.1| Unknown protein {Arabidopsis
thaliana}, partial (88%)
Length = 1029
Score = 452 bits (1162), Expect = e-128
Identities = 226/226 (100%), Positives = 226/226 (100%)
Frame = +1
Query: 1 MGSEGPSAAVTTVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTA 60
MGSEGPSAAVTTVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTA
Sbjct: 181 MGSEGPSAAVTTVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTA 360
Query: 61 GQGALVPLYQTLQSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDE 120
GQGALVPLYQTLQSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDE
Sbjct: 361 GQGALVPLYQTLQSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDE 540
Query: 121 MGWKPQKVPIVPSDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSL 180
MGWKPQKVPIVPSDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSL
Sbjct: 541 MGWKPQKVPIVPSDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSL 720
Query: 181 RFAEQNGNKSLFVHVPLFFTINEETQMQFAASLLEVLANVSNTQLM 226
RFAEQNGNKSLFVHVPLFFTINEETQMQFAASLLEVLANVSNTQLM
Sbjct: 721 RFAEQNGNKSLFVHVPLFFTINEETQMQFAASLLEVLANVSNTQLM 858
>BQ155063 weakly similar to GP|12039349|gb putative pyrrolidone carboxyl
peptidase {Oryza sativa}, partial (65%)
Length = 641
Score = 317 bits (813), Expect = 2e-87
Identities = 172/223 (77%), Positives = 172/223 (77%)
Frame = +3
Query: 1 MGSEGPSAAVTTVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTA 60
MGSEGPSAAVTTVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTA
Sbjct: 126 MGSEGPSAAVTTVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTA 305
Query: 61 GQGALVPLYQTLQSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDE 120
GQGALVPLYQTLQSAIIAKESESSSSNKIIW
Sbjct: 306 GQGALVPLYQTLQSAIIAKESESSSSNKIIW----------------------------- 398
Query: 121 MGWKPQKVPIVPSDGPISRIRETTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSL 180
TTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSL
Sbjct: 399 ----------------------TTLPVEEITKTLATKGYDVMTSDDAGRFVCNYVYYHSL 512
Query: 181 RFAEQNGNKSLFVHVPLFFTINEETQMQFAASLLEVLANVSNT 223
RFAEQNGNKSLFVHVPLFFTINEETQMQFAASLLEVLANVSNT
Sbjct: 513 RFAEQNGNKSLFVHVPLFFTINEETQMQFAASLLEVLANVSNT 641
>TC86515 similar to GP|22136908|gb|AAM91798.1 unknown protein {Arabidopsis
thaliana}, partial (89%)
Length = 1173
Score = 288 bits (737), Expect = 1e-78
Identities = 144/220 (65%), Positives = 174/220 (78%), Gaps = 1/220 (0%)
Frame = +2
Query: 1 MGSEGPSAAVTTVYITGFKKFHGVSENPTETIVNNLTEYVKKKGLPKGLAIGSCSILDTA 60
MGSEGP T+++TGFKKF GV NPTE IVNNL +YV+KKG P G+ +GSC+IL+ A
Sbjct: 101 MGSEGPKKI--TIHVTGFKKFQGVPINPTEIIVNNLRDYVEKKGFPAGVTLGSCTILEVA 274
Query: 61 GQGALVPLYQTLQSAIIAKESESSSSNKIIWLHFGVNSGATRFAIERQAVNEATFRCPDE 120
G AL LYQT++S + ++ES+++ ++WLH GVNSGA RFAIER A NEATFRC DE
Sbjct: 275 GDDALPQLYQTMESVVSKTDTESNAN--LVWLHLGVNSGAARFAIERLAANEATFRCSDE 448
Query: 121 MGWKPQKVPIVPSDGPISRIRETTLPVEEITKTLAT-KGYDVMTSDDAGRFVCNYVYYHS 179
+GW+PQ+VPIV DG ISR RET+LPV+ I K L K YDVM S+DAGRFVCNYVYYHS
Sbjct: 449 LGWQPQQVPIVLEDGGISRTRETSLPVDAILKFLKKGKDYDVMISNDAGRFVCNYVYYHS 628
Query: 180 LRFAEQNGNKSLFVHVPLFFTINEETQMQFAASLLEVLAN 219
LRFAEQ GNKSLFVHVPLF I+EETQM+F +SLLE +A+
Sbjct: 629 LRFAEQKGNKSLFVHVPLFSRIDEETQMRFTSSLLEAIAS 748
>TC80644 similar to GP|20259627|gb|AAM14170.1 unknown protein {Arabidopsis
thaliana}, partial (52%)
Length = 774
Score = 28.5 bits (62), Expect = 2.2
Identities = 12/32 (37%), Positives = 21/32 (65%)
Frame = -1
Query: 56 ILDTAGQGALVPLYQTLQSAIIAKESESSSSN 87
+LDT + +L+ L +Q II K++++ SSN
Sbjct: 672 LLDTQNERSLITLIMIMQMIIIIKQNQNQSSN 577
>TC86664 similar to GP|20340237|gb|AAM19705.1 protein phosphatase 2c-like
protein {Thellungiella halophila}, partial (62%)
Length = 1066
Score = 28.1 bits (61), Expect = 2.9
Identities = 11/35 (31%), Positives = 21/35 (59%)
Frame = +2
Query: 159 YDVMTSDDAGRFVCNYVYYHSLRFAEQNGNKSLFV 193
YD + RF+C++++ H RFA ++ + S+ V
Sbjct: 578 YDGHGGPETSRFICDHLFQHLKRFATEHKSMSVEV 682
>TC85970 homologue to PIR|T50532|T50532 ribosomal protein S27 - Arabidopsis
thaliana, complete
Length = 680
Score = 27.7 bits (60), Expect = 3.7
Identities = 21/71 (29%), Positives = 34/71 (47%)
Frame = -2
Query: 149 EITKTLATKGYDVMTSDDAGRFVCNYVYYHSLRFAEQNGNKSLFVHVPLFFTINEETQMQ 208
+I + TK ++M++ NY+ YH L+F +QN NKS+ F Q +
Sbjct: 547 QINYQI*TKSNEIMST--------NYIRYHLLKFFQQN-NKSIDHKTTRF-------QQR 416
Query: 209 FAASLLEVLAN 219
F +E L+N
Sbjct: 415 FKIQNIETLSN 383
>TC88324 similar to PIR|F86238|F86238 protein T10O24.11 [imported] -
Arabidopsis thaliana, partial (95%)
Length = 815
Score = 27.7 bits (60), Expect = 3.7
Identities = 14/40 (35%), Positives = 21/40 (52%)
Frame = +3
Query: 170 FVCNYVYYHSLRFAEQNGNKSLFVHVPLFFTINEETQMQF 209
F+CNY YY +L G++S ++ LF N +Q F
Sbjct: 63 FLCNYYYYRAL--LSPIGSRSNHSYLSLFSIFNTNSQSPF 176
>BI309682 weakly similar to GP|14532612|g unknown protein {Arabidopsis
thaliana}, partial (16%)
Length = 805
Score = 26.9 bits (58), Expect = 6.4
Identities = 19/60 (31%), Positives = 28/60 (46%), Gaps = 1/60 (1%)
Frame = -1
Query: 164 SDDAGRFVCNYVYYHSLRFAEQNGNK-SLFVHVPLFFTINEETQMQFAASLLEVLANVSN 222
S + FVC Y YHSL EQ K S + +P ++ T+ F S +V N ++
Sbjct: 535 SSGSRSFVCAYKAYHSLAICEQYPTKNSTSMKIPNLVSV--LTEKSFFNSTKQVAKNATS 362
>TC77488 similar to GP|15912313|gb|AAL08290.1 AT5g24690/MXC17_8 {Arabidopsis
thaliana}, partial (79%)
Length = 2036
Score = 26.9 bits (58), Expect = 6.4
Identities = 16/50 (32%), Positives = 25/50 (50%), Gaps = 4/50 (8%)
Frame = -3
Query: 144 TLPVEEITKTLATKGYDVMTSDDAG----RFVCNYVYYHSLRFAEQNGNK 189
T+ + IT T+ ++ T+ +G R V + Y LRF E+NG K
Sbjct: 609 TIIITSITTTITDYLSNISTTSPSG*LRRRLVTLLLRYFKLRFLERNGTK 460
>TC80089
Length = 1618
Score = 26.6 bits (57), Expect = 8.3
Identities = 16/35 (45%), Positives = 20/35 (56%)
Frame = +2
Query: 166 DAGRFVCNYVYYHSLRFAEQNGNKSLFVHVPLFFT 200
D + V N Y+ ++ A QN NKSL H PLF T
Sbjct: 767 DCSQLVSNSGYFGTI--A*QNLNKSLLSHNPLFKT 865
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.132 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,457,893
Number of Sequences: 36976
Number of extensions: 82599
Number of successful extensions: 364
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 360
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 362
length of query: 226
length of database: 9,014,727
effective HSP length: 93
effective length of query: 133
effective length of database: 5,575,959
effective search space: 741602547
effective search space used: 741602547
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC147011.8


