

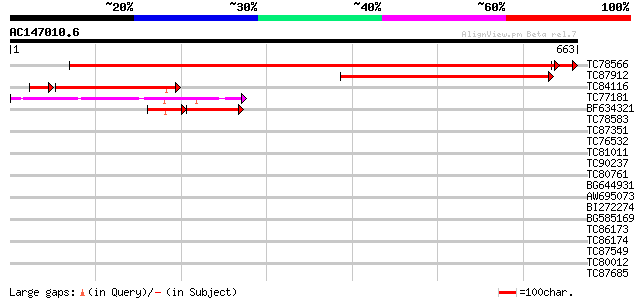
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147010.6 + phase: 0
(663 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78566 similar to GP|20466388|gb|AAM20511.1 putative chloroplas... 1142 0.0
TC87912 homologue to GP|576509|gb|AAA53276.1|| GTP-binding prote... 258 4e-69
TC84116 homologue to PIR|S49910|S49910 chloroplast outer envelop... 199 1e-53
TC77181 similar to GP|510190|emb|CAA82196.1| chloroplast outer e... 130 2e-30
BF634321 weakly similar to GP|576509|gb|A GTP-binding protein {P... 59 2e-19
TC78583 similar to GP|4097585|gb|AAD09518.1| NTGP4 {Nicotiana ta... 40 0.002
TC87351 similar to GP|10176988|dbj|BAB10220. GTP-binding protein... 35 0.093
TC76532 weakly similar to SP|Q9BBN6|YCF1_LOTJA Hypothetical 214.... 35 0.12
TC81011 similar to GP|8489806|gb|AAF75761.1| chloroplast protein... 34 0.16
TC90237 similar to GP|20466544|gb|AAM20589.1 unknown protein {Ar... 33 0.46
TC80761 homologue to GP|1370174|emb|CAA98164.1 RAB1Y {Lotus japo... 32 0.60
BG644931 similar to GP|22135878|gb GTP-binding protein putative... 30 2.3
AW695073 similar to GP|22831198|db putative chromodomain-helicas... 30 2.3
BI272274 similar to PIR|T06443|T064 GTP-binding protein - garden... 30 2.3
BG585169 similar to GP|1370152|emb RAB11F {Lotus japonicus}, par... 30 3.0
TC86173 similar to GP|15810175|gb|AAL06989.1 At2g03890/T18C20.9 ... 30 3.0
TC86174 homologue to OMNI|SO2899 cysZ protein {Shewanella oneide... 30 3.0
TC87549 similar to PIR|T01734|T01734 hypothetical protein A_IG00... 30 3.9
TC80012 GP|2598571|emb|CAA75573.1 putative {Medicago truncatula}... 30 3.9
TC87685 weakly similar to PIR|T02289|T02289 probable polygalactu... 30 3.9
>TC78566 similar to GP|20466388|gb|AAM20511.1 putative chloroplast outer
membrane protein {Arabidopsis thaliana}, partial (46%)
Length = 2245
Score = 1142 bits (2955), Expect(2) = 0.0
Identities = 572/573 (99%), Positives = 573/573 (99%)
Frame = +3
Query: 70 DTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPLLRTITDIF 129
DTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPLLRTITDIF
Sbjct: 3 DTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMPLLRTITDIF 182
Query: 130 GPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDMRLMNPVSL 189
GPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDMRLMNPVSL
Sbjct: 183 GPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAGDMRLMNPVSL 362
Query: 190 VENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQDNPREKPYTARAR 249
VENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQDNPREKPYTARAR
Sbjct: 363 VENHSACRTNTAGQRVLPNGQVWKPQLLLLSFASKILAEANALLKLQDNPREKPYTARAR 542
Query: 250 APPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDPDDLPPFKPLTKAQ 309
APPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDPDDLPPFKPLTKAQ
Sbjct: 543 APPLPFLLSSLLQSRPQLKLPEDQFSDEDSLNDDLDEPSDSGDETDPDDLPPFKPLTKAQ 722
Query: 310 IRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLPSDYVENVEEES 369
IRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLPSDYVENVEEES
Sbjct: 723 IRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLPSDYVENVEEES 902
Query: 370 GGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVLETHGWDHDVGYEGLNVE 429
GGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVL+THGWDHDVGYEGLNVE
Sbjct: 903 GGAASVPVPMPDMSLPASFDSDTPTHRYRHLDSSNQWLVRPVLKTHGWDHDVGYEGLNVE 1082
Query: 430 RLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATSLGFDMQTVGKDLAYTLR 489
RLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATSLGFDMQTVGKDLAYTLR
Sbjct: 1083RLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVKYGEGKATSLGFDMQTVGKDLAYTLR 1262
Query: 490 SETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAGGAMTGRDDVAYG 549
SETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAGGAMTGRDDVAYG
Sbjct: 1263SETKFCNFLRNKATAGLSFTLLGDALSAGVKVEDKLIANKRFKLVIAGGAMTGRDDVAYG 1442
Query: 550 GSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYTNLVARANLNNRG 609
GSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYTNLVARANLNNRG
Sbjct: 1443GSLEAQLRDKNYPLGRSLSTLGLSVMDWHGDLAVGCNLQSQIPIGRYTNLVARANLNNRG 1622
Query: 610 AGQISIRLNSSEQLQIALIGLIPLLKKVIGYSQ 642
AGQISIRLNSSEQLQIALIGLIPLLKKVIGYSQ
Sbjct: 1623AGQISIRLNSSEQLQIALIGLIPLLKKVIGYSQ 1721
Score = 50.1 bits (118), Expect(2) = 0.0
Identities = 22/30 (73%), Positives = 27/30 (89%)
Frame = +1
Query: 634 LKKVIGYSQNCSLDKIIDEVRLCEMHNSSR 663
L++++ +NCSLDKIIDEVRLCEMHNSSR
Sbjct: 1696 LRRLLVILKNCSLDKIIDEVRLCEMHNSSR 1785
>TC87912 homologue to GP|576509|gb|AAA53276.1|| GTP-binding protein {Pisum
sativum}, partial (30%)
Length = 1126
Score = 258 bits (660), Expect = 4e-69
Identities = 119/248 (47%), Positives = 177/248 (70%)
Frame = +2
Query: 388 FDSDTPTHRYRHLDSSNQWLVRPVLETHGWDHDVGYEGLNVERLFVLKDKIPVSFSGQVT 447
FDSD P +RYR L+ ++Q L RPVL+TH WDHD GY+G+N+E + +K P + + QVT
Sbjct: 2 FDSDNPAYRYRFLEPTSQLLTRPVLDTHSWDHDCGYDGVNIENSVAIINKFPAAVTVQVT 181
Query: 448 KDKKDANVQMEMTSSVKYGEGKATSLGFDMQTVGKDLAYTLRSETKFCNFLRNKATAGLS 507
KDK+D ++ ++ + + K+GE +T GFD+Q +GK +AY +R ETKF NF RNK AG+S
Sbjct: 182 KDKQDFSIHLDSSVAAKHGENGSTMAGFDIQNIGKQMAYIVRGETKFKNFKRNKTAAGVS 361
Query: 508 FTLLGDALSAGVKVEDKLIANKRFKLVIAGGAMTGRDDVAYGGSLEAQLRDKNYPLGRSL 567
T LG+ +S GVK+ED+L KR LV + G + + D AYG ++E +LR+ ++P+G+
Sbjct: 362 VTFLGENVSTGVKLEDQLALGKRLVLVGSTGTVRSQGDSAYGANVEVRLREADFPIGQDQ 541
Query: 568 STLGLSVMDWHGDLAVGCNLQSQIPIGRYTNLVARANLNNRGAGQISIRLNSSEQLQIAL 627
S+L S++ W GDLA+G N QSQI +GR + RA LNN+ +GQI++R +SS+QLQIAL
Sbjct: 542 SSLSFSLVQWRGDLALGANFQSQISLGRSYKMAVRAGLNNKLSGQITVRTSSSDQLQIAL 721
Query: 628 IGLIPLLK 635
I ++P+++
Sbjct: 722 IAMLPIVR 745
>TC84116 homologue to PIR|S49910|S49910 chloroplast outer envelope protein
OEP86 precursor - garden pea, partial (20%)
Length = 555
Score = 199 bits (505), Expect(2) = 1e-53
Identities = 94/151 (62%), Positives = 118/151 (77%), Gaps = 5/151 (3%)
Frame = +2
Query: 54 VQDVVGMVQGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQS 113
V ++VGMV G+KVRV DTPGL S +Q +N K+L +VK+ K +PPDIVLY+DRLD+Q+
Sbjct: 95 VTEIVGMVDGVKVRVFDTPGLKSSAFEQSYNRKVLSNVKKLTKNSPPDIVLYVDRLDLQT 274
Query: 114 RDFSDMPLLRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQA 173
RD +D+P+LR++T GP IW N IV LTHAASAPPDGP+G+P SYD+FV QR+H+VQQ
Sbjct: 275 RDMNDLPMLRSVTTALGPSIWRNVIVTLTHAASAPPDGPSGSPLSYDVFVAQRTHIVQQT 454
Query: 174 IRQAAGDMR-----LMNPVSLVENHSACRTN 199
I QA GD+R LMNPVSLVENH +CR N
Sbjct: 455 IGQAVGDLRLMNPSLMNPVSLVENHPSCRKN 547
Score = 30.0 bits (66), Expect(2) = 1e-53
Identities = 15/28 (53%), Positives = 18/28 (63%)
Frame = +1
Query: 24 KSGVGKSSTINSIFDEVKFNTDAFHMGT 51
K VGKS+TINSIF E K + A+ T
Sbjct: 4 KLAVGKSATINSIFGETKTSFSAYGPAT 87
>TC77181 similar to GP|510190|emb|CAA82196.1| chloroplast outer envelope
protein 34 {Pisum sativum}, partial (96%)
Length = 1118
Score = 130 bits (326), Expect = 2e-30
Identities = 98/288 (34%), Positives = 137/288 (47%), Gaps = 11/288 (3%)
Frame = +2
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGM 60
+ E L QE ++ S TI+V+GK GVGKSST+NSI E F + V
Sbjct: 239 LLELLGKLKQEDVN-SLTILVMGKGGVGKSSTVNSIIGERVVAISPFQSEGPRPVMVSRA 415
Query: 61 VQGIKVRVIDTPGLLPSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMP 120
G + +IDTPGL+ N+ L +KRF+ D++LY+DRLD D D
Sbjct: 416 RAGFTLNIIDTPGLIEGGYI---NDMALDIIKRFLLDKTIDVLLYVDRLDAYRVDNLDKL 586
Query: 121 LLRTITDIFGPPIWFNAIVVLTHAASAPPDGPNGTPSSYDMFVTQRSHVVQQAIRQAAG- 179
+ + ITD FG IW AIV LTHA +PPD +YD F ++RS + Q I+ A
Sbjct: 587 VAKAITDSFGKGIWNKAIVALTHAQFSPPDA-----LAYDEFFSKRSESLLQIIKSGASL 751
Query: 180 ----DMRLMNPVSLVENHSACRTNTAGQRVLPNGQVWKPQL------LLLSFASKILAEA 229
PV L+EN C N ++VLPNG W P L + L+ + I +
Sbjct: 752 KKDDAQASAIPVVLIENSGRCNKNETDEKVLPNGIAWIPHLVHTITEIALNKSESIFVDK 931
Query: 230 NALLKLQDNPREKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQFSDE 277
N + N R K + P+ F L LL +P L ++ ++E
Sbjct: 932 NLIEGPNPNQRGKLWI------PVLFALEFLLIMKPIEGLIKNDVANE 1057
>BF634321 weakly similar to GP|576509|gb|A GTP-binding protein {Pisum
sativum}, partial (12%)
Length = 384
Score = 58.9 bits (141), Expect(2) = 2e-19
Identities = 32/68 (47%), Positives = 43/68 (63%), Gaps = 1/68 (1%)
Frame = +3
Query: 207 PNGQVWKPQLLLLSFASKILAEANALLKLQDN-PREKPYTARARAPPLPFLLSSLLQSRP 265
PNGQ W+P LLLL ++ KIL+EA +L K Q+ K + R + P LP+LLS L+Q R
Sbjct: 153 PNGQTWRPLLLLLCYSMKILSEATSLSKTQEMFNYNKLFGFRVQTPSLPYLLSWLMQPRN 332
Query: 266 QLKLPEDQ 273
KL +Q
Sbjct: 333 HAKLASNQ 356
Score = 55.5 bits (132), Expect(2) = 2e-19
Identities = 29/50 (58%), Positives = 34/50 (68%), Gaps = 5/50 (10%)
Frame = +2
Query: 162 FVTQRSHVVQQAIRQAAGD-----MRLMNPVSLVENHSACRTNTAGQRVL 206
FVTQR+ VQQAI Q GD + LMNPV+LVENH +CR N G +VL
Sbjct: 2 FVTQRNRAVQQAIGQVIGDEQINNLSLMNPVALVENHFSCRKNKNGHKVL 151
>TC78583 similar to GP|4097585|gb|AAD09518.1| NTGP4 {Nicotiana tabacum},
partial (86%)
Length = 1269
Score = 40.4 bits (93), Expect = 0.002
Identities = 41/129 (31%), Positives = 61/129 (46%), Gaps = 4/129 (3%)
Frame = +3
Query: 18 TIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKK--VQDVVGMVQGIKVRVIDTPGLL 75
T++++G++G GKS+T NSI + F + A G + G V VIDTPGL
Sbjct: 138 TVVLVGRTGNGKSATGNSILGKKVFKSRASSSGVTSSCEMQTAELSDGQIVNVIDTPGL- 314
Query: 76 PSWSDQPHNEKILHSVKRFIKKTPPDIVLYLDRLDMQSRDFSDMP--LLRTITDIFGPPI 133
+ +E I + + I I L L ++SR FS+ LR++ +FG I
Sbjct: 315 --FEVSAGSEFIGKEIVKCIDFAKDGIHAILVVLSVRSR-FSEEEENALRSLQTLFGSKI 485
Query: 134 WFNAIVVLT 142
IVV T
Sbjct: 486 VDYMIVVFT 512
>TC87351 similar to GP|10176988|dbj|BAB10220. GTP-binding protein-like
{Arabidopsis thaliana}, partial (81%)
Length = 2266
Score = 35.0 bits (79), Expect = 0.093
Identities = 18/60 (30%), Positives = 37/60 (61%), Gaps = 3/60 (5%)
Frame = +3
Query: 18 TIMVLGKSGVGKSSTINSIF---DEVKFNTDAFHMGTKKVQDVVGMVQGIKVRVIDTPGL 74
T+++LG+ VGKS+ N + + + +NT H+ T+ +++ V + ++ RV+D+ GL
Sbjct: 363 TVIILGRPNVGKSALFNRLIRRREALVYNTPDDHV-TRDIREGVAKLGHLRFRVLDSAGL 539
>TC76532 weakly similar to SP|Q9BBN6|YCF1_LOTJA Hypothetical 214.8 kDa protein
ycf1. {Lotus japonicus}, partial (21%)
Length = 4509
Score = 34.7 bits (78), Expect = 0.12
Identities = 27/110 (24%), Positives = 58/110 (52%), Gaps = 3/110 (2%)
Frame = +1
Query: 310 IRNLSRAQKKAYLDEVEYREKLFMKKQLKYEKKQRKMMKEMAESVKDLPSDYVENVEEES 369
I +++R ++ +LD +E +K+ K+ Y+KK K M+EM E ++L + ++ E+S
Sbjct: 2332 ISSIARIHRQRFLDFLESTDKILNVKKPIYDKK--KKMEEMEERFENLSVSRLISILEKS 2505
Query: 370 GGAASVPVPMP-DMSLPASFDSDTPTHRYRHLDSSN--QWLVRPVLETHG 416
++ D+S +S + ++ + SN ++ +R +LE+ G
Sbjct: 2506 ENITNMNSQNSWDVSSLSSLSQEYVFYKLSQIQFSNGSKFKIRSILESPG 2655
>TC81011 similar to GP|8489806|gb|AAF75761.1| chloroplast protein import
component Toc159 {Pisum sativum}, partial (17%)
Length = 808
Score = 34.3 bits (77), Expect = 0.16
Identities = 16/23 (69%), Positives = 19/23 (82%)
Frame = +1
Query: 15 FSCTIMVLGKSGVGKSSTINSIF 37
FS I+VLGK+GVGKS+TIN F
Sbjct: 727 FSINILVLGKTGVGKSATINFNF 795
>TC90237 similar to GP|20466544|gb|AAM20589.1 unknown protein {Arabidopsis
thaliana}, partial (79%)
Length = 1174
Score = 32.7 bits (73), Expect = 0.46
Identities = 29/89 (32%), Positives = 42/89 (46%), Gaps = 14/89 (15%)
Frame = +1
Query: 6 ESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQGI- 64
E+ +EP + +MV+G VGKS INSI + F + K + VG + G+
Sbjct: 307 EAISKEP---TLLVMVVGVPNVGKSCLINSIH---QIAHSRFPVQEKMKRAAVGPLPGVT 468
Query: 65 ------------KVRVIDTPG-LLPSWSD 80
+ V+DTPG L+PS SD
Sbjct: 469 QDIAGFKIANKPSIYVLDTPGVLVPSISD 555
>TC80761 homologue to GP|1370174|emb|CAA98164.1 RAB1Y {Lotus japonicus},
partial (94%)
Length = 994
Score = 32.3 bits (72), Expect = 0.60
Identities = 23/73 (31%), Positives = 40/73 (54%), Gaps = 6/73 (8%)
Frame = +1
Query: 7 SAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFH-----MGTK-KVQDVVGM 60
S+GQ+ D+ ++++G SGVGKSS + ++F +D F +G KV+ V
Sbjct: 121 SSGQQEFDYMFKLLMIGDSGVGKSSLL------LRFTSDDFDDLSPTIGVDFKVKYVTIE 282
Query: 61 VQGIKVRVIDTPG 73
+ +K+ + DT G
Sbjct: 283 GKKLKLAIWDTAG 321
>BG644931 similar to GP|22135878|gb GTP-binding protein putative
{Arabidopsis thaliana}, partial (47%)
Length = 697
Score = 30.4 bits (67), Expect = 2.3
Identities = 34/130 (26%), Positives = 56/130 (42%), Gaps = 9/130 (6%)
Frame = +3
Query: 18 TIMVLGKSGVGKSSTINSIFD------EVKFNTDAFHMGTKKVQDVVGMVQGIKVRVIDT 71
T+ ++G VGKSS ++ + F T MG +V Q K +V DT
Sbjct: 174 TLCLVGAPNVGKSSLVHVLSTGKPEICNYPFTTRGILMG-----HIVFNHQ--KFQVTDT 332
Query: 72 PGLLPSWSDQPHN-EKILHSVKRFIKKTPPDIVLYLDRL--DMQSRDFSDMPLLRTITDI 128
PGLL D +N EK+ +V ++ P VLY+ L + + + + I +
Sbjct: 333 PGLLKRHDDDRNNLEKLTLAVLSYL----PTAVLYVHDLSGECGTSPSDQYSIYKEIKER 500
Query: 129 FGPPIWFNAI 138
F +W + +
Sbjct: 501 FDGHLWLDVV 530
>AW695073 similar to GP|22831198|db putative
chromodomain-helicase-DNA-binding protein {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 449
Score = 30.4 bits (67), Expect = 2.3
Identities = 18/51 (35%), Positives = 25/51 (48%)
Frame = +2
Query: 276 DEDSLNDDLDEPSDSGDETDPDDLPPFKPLTKAQIRNLSRAQKKAYLDEVE 326
D + +DD D D DE DPDD F+P T + N + + DEV+
Sbjct: 218 DAEDDDDDADYEEDEPDEDDPDD-ADFEPATSGRGANKYKDWEGEDSDEVD 367
>BI272274 similar to PIR|T06443|T064 GTP-binding protein - garden pea,
partial (32%)
Length = 689
Score = 30.4 bits (67), Expect = 2.3
Identities = 16/52 (30%), Positives = 28/52 (53%)
Frame = +3
Query: 1 MAEQLESAGQEPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTK 52
MA+ E +D+ ++++G SGVGKS +N +F + FH+ +K
Sbjct: 321 MAQFQGDLDHEGIDYMFKVVMIGDSGVGKSQLLN------RFVRNEFHLKSK 458
>BG585169 similar to GP|1370152|emb RAB11F {Lotus japonicus}, partial (83%)
Length = 668
Score = 30.0 bits (66), Expect = 3.0
Identities = 19/57 (33%), Positives = 29/57 (50%)
Frame = +3
Query: 11 EPLDFSCTIMVLGKSGVGKSSTINSIFDEVKFNTDAFHMGTKKVQDVVGMVQGIKVR 67
E D+ +++G SGVGKS+ I+ +F D F + +K V + IKVR
Sbjct: 138 EQCDYLFKTVLIGDSGVGKSNLIS------RFTKDEFRLDSKPTIGVEFGYKNIKVR 290
>TC86173 similar to GP|15810175|gb|AAL06989.1 At2g03890/T18C20.9
{Arabidopsis thaliana}, partial (74%)
Length = 2669
Score = 30.0 bits (66), Expect = 3.0
Identities = 15/44 (34%), Positives = 24/44 (54%)
Frame = +2
Query: 602 RANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKVIGYSQNCS 645
R N R +IS L + L +L+ +PL ++ GYS++CS
Sbjct: 23 RRNTRARFVTEISSELTRTSLLYDSLLSFLPLSEQFDGYSEDCS 154
>TC86174 homologue to OMNI|SO2899 cysZ protein {Shewanella oneidensis MR-1},
partial (4%)
Length = 642
Score = 30.0 bits (66), Expect = 3.0
Identities = 15/44 (34%), Positives = 24/44 (54%)
Frame = -2
Query: 602 RANLNNRGAGQISIRLNSSEQLQIALIGLIPLLKKVIGYSQNCS 645
R N R +IS L + L +L+ +PL ++ GYS++CS
Sbjct: 626 RRNTRARFVTEISSELTRTSLLYDSLLSFLPLSEQFDGYSEDCS 495
>TC87549 similar to PIR|T01734|T01734 hypothetical protein A_IG002N01.30 -
Arabidopsis thaliana (fragment), partial (38%)
Length = 1766
Score = 29.6 bits (65), Expect = 3.9
Identities = 24/78 (30%), Positives = 33/78 (41%)
Frame = +2
Query: 271 EDQFSDEDSLNDDLDEPSDSGDETDPDDLPPFKPLTKAQIRNLSRAQKKAYLDEVEYREK 330
+D SD+D +DD+D+ D DE D D P F DE +
Sbjct: 1577 DDLISDDDDDDDDIDD--DWIDEDDDDSPPDFD-------------------DEEAETSE 1693
Query: 331 LFMKKQLKYEKKQRKMMK 348
L K+QL K Q ++MK
Sbjct: 1694 LKRKRQLXXFKTQDRIMK 1747
>TC80012 GP|2598571|emb|CAA75573.1 putative {Medicago truncatula}, complete
Length = 511
Score = 29.6 bits (65), Expect = 3.9
Identities = 18/43 (41%), Positives = 23/43 (52%)
Frame = -1
Query: 232 LLKLQDNPREKPYTARARAPPLPFLLSSLLQSRPQLKLPEDQF 274
L +L P KP + R PLPFLL+SL P++ L QF
Sbjct: 127 LPELYSVPPPKPASFR----PLPFLLASLFLPSPKIPLSSSQF 11
>TC87685 weakly similar to PIR|T02289|T02289 probable polygalacturonase (EC
3.2.1.15) 1 beta chain T13D8.26 - Arabidopsis thaliana,
partial (42%)
Length = 1572
Score = 29.6 bits (65), Expect = 3.9
Identities = 15/47 (31%), Positives = 20/47 (41%)
Frame = +1
Query: 418 DHDVGYEGLNVERLFVLKDKIPVSFSGQVTKDKKDANVQMEMTSSVK 464
DH + ++G V F DK +SF+G K N E S K
Sbjct: 1135 DHKITFKGYGVNNTFKDYDKKGISFAGYTKKSSSSTNSVSESVSLAK 1275
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,948,969
Number of Sequences: 36976
Number of extensions: 232354
Number of successful extensions: 1390
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 1344
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1379
length of query: 663
length of database: 9,014,727
effective HSP length: 102
effective length of query: 561
effective length of database: 5,243,175
effective search space: 2941421175
effective search space used: 2941421175
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147010.6


