

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147010.5 - phase: 0
(657 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
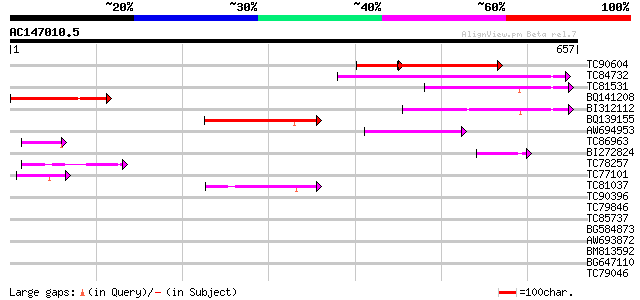
Score E
Sequences producing significant alignments: (bits) Value
TC90604 weakly similar to PIR|B96760|B96760 probable homeobox pr... 219 2e-75
TC84732 homologue to GP|14334996|gb|AAK59762.1 At1g05230/YUP8H12... 168 7e-42
TC81531 homologue to GP|3925363|gb|AAC79430.1| homeodomain prote... 115 5e-26
BQ141208 similar to PIR|S71477|S71 homeotic protein ovule-speci... 113 2e-25
BI312112 similar to GP|22475195|gb homeodomain protein GhHOX1 {G... 112 5e-25
BQ139155 similar to GP|3925363|gb| homeodomain protein {Malus x ... 111 8e-25
AW694953 similar to GP|14334996|gb At1g05230/YUP8H12_16 {Arabido... 86 5e-17
TC86963 similar to GP|15148918|gb|AAK84886.1 homeodomain leucine... 46 5e-05
BI272824 similar to GP|1814424|gb|A homeodomain protein AHDP {Ar... 45 1e-04
TC78257 similar to PIR|T47981|T47981 homeobox-leucine zipper pro... 43 3e-04
TC77101 similar to GP|15148920|gb|AAK84887.1 homeodomain leucine... 42 6e-04
TC81037 homologue to GP|18076740|emb|CAC84277. HD-Zip protein {Z... 42 7e-04
TC90396 similar to PIR|H96719|H96719 homeobox gene 13 protein 1... 42 0.001
TC79846 similar to GP|8843728|dbj|BAA97276.1 homeodomain transcr... 42 0.001
TC85737 similar to GP|6091551|gb|AAF01764.2| homeodomain-leucine... 41 0.002
BG584873 similar to GP|20161555|dbj putative homeodomain-leucine... 40 0.002
AW693872 similar to PIR|D84679|D846 probable homeodomain transcr... 40 0.002
BM813592 similar to GP|20161555|db putative homeodomain-leucine ... 40 0.002
BG647110 homologue to PIR|T10695|T10 transcription factor HD-zip... 40 0.003
TC79046 similar to PIR|T12634|T12634 homeotic protein - common s... 39 0.006
>TC90604 weakly similar to PIR|B96760|B96760 probable homeobox protein
T9L24.43 [imported] - Arabidopsis thaliana, partial (8%)
Length = 1097
Score = 219 bits (558), Expect(2) = 2e-75
Identities = 105/122 (86%), Positives = 112/122 (91%)
Frame = +1
Query: 450 LASIGGIRVSFRSTTDDDTSQPNGTIVTAATTLWLPLPALKVFEFLKDPTKRSQWDGLSC 509
L ++ G RVS R TTD+DTSQPNGT+VTAATTLWLPLPA KVFE LKDPTKRSQWDGLSC
Sbjct: 562 LIALVGFRVSLRDTTDNDTSQPNGTVVTAATTLWLPLPAQKVFELLKDPTKRSQWDGLSC 741
Query: 510 GNPMHEIAHISNGPYHGNCISIIKPFIPTQRQMMILQESFTSRVGSYIIYAPSDRQTMDV 569
GNPMHEIAHISNGPYHGNCISIIK FIPTQRQM+ILQESFTS VGSYIIYAP DR+T+DV
Sbjct: 742 GNPMHEIAHISNGPYHGNCISIIKSFIPTQRQMVILQESFTSPVGSYIIYAPIDRKTVDV 921
Query: 570 AL 571
AL
Sbjct: 922 AL 927
Score = 82.4 bits (202), Expect(2) = 2e-75
Identities = 42/55 (76%), Positives = 45/55 (81%)
Frame = +3
Query: 402 EAIPVEETIGVIQTLEGRNSVIKLAQRMVKMFCESLTMPGQLELNHLTLASIGGI 456
EA E +IQTLEGRNSVIKLA RMVKMFCE LTMPGQ+ELNHLTL SIGG+
Sbjct: 417 EAEEATEERRLIQTLEGRNSVIKLADRMVKMFCECLTMPGQVELNHLTLDSIGGV 581
>TC84732 homologue to GP|14334996|gb|AAK59762.1 At1g05230/YUP8H12_16
{Arabidopsis thaliana}, partial (35%)
Length = 808
Score = 168 bits (425), Expect = 7e-42
Identities = 101/272 (37%), Positives = 147/272 (53%), Gaps = 2/272 (0%)
Frame = +3
Query: 381 GAESWIKELQRMCERSLGSYVEAIPVEETIGVIQTLEGRNSVIKLAQRMVKMFCESLTMP 440
GA+ WI L R CER + IP + +GVI +GR S++KLA+RM FC ++
Sbjct: 3 GAKRWIATLDRQCERLASAMATNIPTVD-VGVITNQDGRKSMLKLAERMCISFCAGVSAS 179
Query: 441 GQLELNHLTLASIGGIRVSFRSTTDDDTSQPNGTIVTAATTLWLPLPALKVFEFLKDPTK 500
L+ +RV R + DD +P G +++AAT+ WLP+P +VFEFL+D
Sbjct: 180 TAHTWTTLSGTGADDVRVMTRKSVDDP-GRPAGIVLSAATSFWLPVPPKRVFEFLRDENS 356
Query: 501 RSQWDGLSCGNPMHEIAHISNGPYHGNCISIIK--PFIPTQRQMMILQESFTSRVGSYII 558
RS+WD LS G + E+AHI+NG GNC+S+++ +Q M+ILQES T GS++I
Sbjct: 357 RSEWDILSNGGVVQEMAHIANGRDTGNCVSLLRVNSANSSQSNMLILQESCTDTTGSFVI 536
Query: 559 YAPSDRQTMDVALRGEDSKELPILPYGFVVCSKSQPNLNAPFGASNNIEDGSLLTLAAQI 618
YAP D M+V L G D + +LP GF + ++ GSLLT+A QI
Sbjct: 537 YAPVDIVAMNVVLNGGDPDYVALLPSGFAILPDGTTTNGGKVLVKLDMGGGSLLTVALQI 716
Query: 619 LSTSPHEIDQVLNVEDITDINTHLATTILNVK 650
L S L++ + +N +A T +K
Sbjct: 717 LVDSVPTAK--LSLGSVATVNNLIACTGERIK 806
>TC81531 homologue to GP|3925363|gb|AAC79430.1| homeodomain protein {Malus x
domestica}, partial (25%)
Length = 888
Score = 115 bits (288), Expect = 5e-26
Identities = 69/181 (38%), Positives = 100/181 (55%), Gaps = 8/181 (4%)
Frame = +3
Query: 481 TLWLPLPALKVFEFLKDPTKRSQWDGLSCGNPMHEIAHISNGPYHGNCISIIK--PFIPT 538
++WLP+ ++F+FL+D RS+WD LS G PM E+AHI+ G HGNC+S+++
Sbjct: 3 SVWLPVSPQRLFDFLRDERLRSEWDILSNGGPMQEMAHIAKGQDHGNCVSLLRASAMNSN 182
Query: 539 QRQMMILQESFTSRVGSYIIYAPSDRQTMDVALRGEDSKELPILPYGFVVC-----SKSQ 593
Q M+ILQE+ GS ++YAP D M V + G DS + +LP GF + S+
Sbjct: 183 QSSMLILQETCIDEAGSLVVYAPVDIPAMHVVMNGGDSAYVALLPSGFAIVPDGPGSRGL 362
Query: 594 PNLNAPF-GASNNIEDGSLLTLAAQILSTSPHEIDQVLNVEDITDINTHLATTILNVKDA 652
N +A G GSLLT+A QIL S L VE + +N ++ T+ +K A
Sbjct: 363 ENGDATANGGGEARVSGSLLTVAFQILVNSLPTAK--LTVESVETVNNLISCTVQKIKAA 536
Query: 653 L 653
L
Sbjct: 537 L 539
>BQ141208 similar to PIR|S71477|S71 homeotic protein ovule-specific -
Phalaenopsis sp., partial (19%)
Length = 661
Score = 113 bits (283), Expect = 2e-25
Identities = 52/117 (44%), Positives = 80/117 (67%)
Frame = +3
Query: 2 KECHHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKIRNENL 61
KEC HPD+ +R +L+ E+GLEP Q+KFWFQNKRT +K QHER N LR +N+K+R +N+
Sbjct: 192 KECPHPDDKQRKELSRELGLEPLQVKFWFQNKRTQMKTQHERSENSQLRADNEKLRADNM 371
Query: 62 KIKEVLKAKICLDCGGPPFPMKDHQNFVQDLKQENAQLKQECEKMSSLLASYMEKKI 118
+ +E L C +CGGP + + L+ ENA+L++E +++S++ A Y+ K +
Sbjct: 372 RYREALSNASCPNCGGPT-AIGEMSFDEHHLRLENARLREEIDRISTMAAKYVGKPV 539
>BI312112 similar to GP|22475195|gb homeodomain protein GhHOX1 {Gossypium
hirsutum}, partial (27%)
Length = 727
Score = 112 bits (280), Expect = 5e-25
Identities = 68/201 (33%), Positives = 114/201 (55%), Gaps = 3/201 (1%)
Frame = +2
Query: 456 IRVSFRSTTDDDTSQPNGTIVTAATTLWLPLPALKVFEFLKDPTKRSQWDGLSCGNPMHE 515
IR+S R +D S+P G IV A +++WLP+ +F+FL+D T+R++WD +S G +
Sbjct: 26 IRISSRKNLNDP-SEPLGLIVCAVSSIWLPISPNVLFDFLRDETRRTEWDIMSNGGTVQS 202
Query: 516 IAHISNGPYHGNCISIIKPFIPTQRQMMILQESFTSRVGSYIIYAPSDRQTMDVALRGED 575
IA+++ G GN ++ I+ + M ILQ+S T+ S ++YAP+D + + G D
Sbjct: 203 IANLAKGQDRGNAVT-IQTIKSKENNMWILQDSCTNSYESMVVYAPADITGIQSVMTGCD 379
Query: 576 SKELPILPYGFVVCS---KSQPNLNAPFGASNNIEDGSLLTLAAQILSTSPHEIDQVLNV 632
S L ILP GF + S +S+ + N E GSL T+A QIL+ + L +
Sbjct: 380 SSNLAILPSGFSIVSDGLESRQMVITSRREEKNTEGGSLFTIAFQILTNASPTAK--LTM 553
Query: 633 EDITDINTHLATTILNVKDAL 653
E + +N+ ++ T+ ++K +L
Sbjct: 554 ESVDSMNSLVSCTLRHIKTSL 616
>BQ139155 similar to GP|3925363|gb| homeodomain protein {Malus x domestica},
partial (24%)
Length = 505
Score = 111 bits (278), Expect = 8e-25
Identities = 55/139 (39%), Positives = 86/139 (61%), Gaps = 3/139 (2%)
Frame = +1
Query: 226 ESSKYSGLVKISGIDLVGMFLDSVKWTNLFPTIVTKAETIKVFEIGSPGSRDGALLLMNE 285
E+S+ SG+V I+ + LV +DS +W+ +FP ++ ++ T +V G G+R+GAL LM
Sbjct: 85 EASRESGVVIINSLALVETLMDSNRWSEMFPCVIARSSTTEVISSGINGTRNGALQLMQA 264
Query: 286 EMHILSPLVRPREFNIIRYCKKFDAGVWVIADVSFDSSRPNTA---PLSRGWKHPSGCII 342
E+ +LSPLV RE + +R+CK+ GVW + DVS D+ +A + PSG ++
Sbjct: 265 ELQVLSPLVPVREVSFLRFCKQHAEGVWAVVDVSIDTIXETSAGAPTFLTCRRLPSGXVV 444
Query: 343 REMPHGGCLVTWVEHVEVE 361
++MP+G VTWVEH E
Sbjct: 445 QDMPNGYSKVTWVEHAXYE 501
>AW694953 similar to GP|14334996|gb At1g05230/YUP8H12_16 {Arabidopsis
thaliana}, partial (16%)
Length = 404
Score = 85.9 bits (211), Expect = 5e-17
Identities = 44/118 (37%), Positives = 70/118 (59%)
Frame = +2
Query: 412 VIQTLEGRNSVIKLAQRMVKMFCESLTMPGQLELNHLTLASIGGIRVSFRSTTDDDTSQP 471
VI +G+ S++KLA+RM FC ++ L+ +RV R + +D +P
Sbjct: 53 VITNQDGKKSMLKLAERMCITFCAGVSASTAHTWTTLSGTGADDVRVMTRKSVNDP-ERP 229
Query: 472 NGTIVTAATTLWLPLPALKVFEFLKDPTKRSQWDGLSCGNPMHEIAHISNGPYHGNCI 529
G +++AAT+ WLP+P +VFEFLK+ +S+WD LS G + ++AHI+NG GN +
Sbjct: 230 AGIVLSAATSFWLPVPPKRVFEFLKNENSKSEWDILSNGGVVQKMAHIANGKNTGNWV 403
>TC86963 similar to GP|15148918|gb|AAK84886.1 homeodomain leucine zipper
protein HDZ2 {Phaseolus vulgaris}, partial (87%)
Length = 1519
Score = 45.8 bits (107), Expect = 5e-05
Identities = 23/59 (38%), Positives = 35/59 (58%), Gaps = 7/59 (11%)
Frame = +1
Query: 14 QLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKIRN-------ENLKIKE 65
QLA E+GL+P+Q+ WFQN+R K + + GTL+ D +++ EN K+KE
Sbjct: 820 QLAKELGLQPRQVAIWFQNRRARFKTKQLEKDYGTLKASFDSLKDDYDNLLQENDKLKE 996
>BI272824 similar to GP|1814424|gb|A homeodomain protein AHDP {Arabidopsis
thaliana}, partial (8%)
Length = 268
Score = 44.7 bits (104), Expect = 1e-04
Identities = 25/65 (38%), Positives = 39/65 (59%), Gaps = 2/65 (3%)
Frame = +1
Query: 542 MMILQESFTSRVGSYIIYAPSDRQTMDVALRGEDSKELPILPYGFVVCSKSQPNLNAP-- 599
M+ILQE++ S ++YAP D Q+++V + G DS + +LP GF + P+ N P
Sbjct: 46 MLILQETWMDTSCSVVVYAPVDGQSLNVVMSGGDSAYVALLPSGFAIV----PDGNDPSG 213
Query: 600 FGASN 604
+G SN
Sbjct: 214 YGMSN 228
>TC78257 similar to PIR|T47981|T47981 homeobox-leucine zipper protein
ATHB-12 - Arabidopsis thaliana, partial (45%)
Length = 994
Score = 43.1 bits (100), Expect = 3e-04
Identities = 35/125 (28%), Positives = 59/125 (47%), Gaps = 2/125 (1%)
Frame = +2
Query: 14 QLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKIRN--ENLKIKEVLKAKI 71
QLA E+GL+P+Q+ WFQNKR K + L RE +K++N NL K
Sbjct: 332 QLARELGLQPRQVAIWFQNKRARWKSKQ-------LEREYNKLQNSYNNLASK------- 469
Query: 72 CLDCGGPPFPMKDHQNFVQDLKQENAQLKQECEKMSSLLASYMEKKISRPEFEQALKSIK 131
+ +K+E L + +K++ L+ +E+ S + ++A KS++
Sbjct: 470 -----------------FESMKKERQTLLIQLQKLNDLIQKPIEQSQSSSQVKEA-KSME 595
Query: 132 SFSRD 136
S S +
Sbjct: 596 SASEN 610
>TC77101 similar to GP|15148920|gb|AAK84887.1 homeodomain leucine zipper
protein HDZ3 {Phaseolus vulgaris}, complete
Length = 1532
Score = 42.4 bits (98), Expect = 6e-04
Identities = 26/77 (33%), Positives = 44/77 (56%), Gaps = 15/77 (19%)
Frame = +1
Query: 9 EAERH-QLAVEVGLEPKQIKFWFQNKRTLLK-HQHERE-------------TNGTLRREN 53
E ER QLA ++GL+P+Q+ WFQN+R K Q ER+ T ++ +EN
Sbjct: 700 EPERKTQLAKKLGLQPRQVAVWFQNRRARWKTKQLERDYDVLKSSYDSLLSTYDSINKEN 879
Query: 54 DKIRNENLKIKEVLKAK 70
+K+++E + + E L+ +
Sbjct: 880 EKLKSEVVSLNEKLQVQ 930
>TC81037 homologue to GP|18076740|emb|CAC84277. HD-Zip protein {Zinnia
elegans}, partial (25%)
Length = 650
Score = 42.0 bits (97), Expect = 7e-04
Identities = 32/140 (22%), Positives = 59/140 (41%), Gaps = 5/140 (3%)
Frame = +1
Query: 227 SSKYSGLVKISGIDLVGMFLDSVKWTNLFPTIVTKAETIKVFEIGSPGSRDGALLLMNEE 286
+++ GLV + + + D + W + ++VF + P G + L+ +
Sbjct: 40 AARACGLVSLEPTKIAEILKDRLSWFR-------ECRNLEVFTM-FPAGNGGTIELIYTQ 195
Query: 287 MHILSPLVRPREFNIIRYCKKFDAGVWVIADVSFDSSRPNTAPLS-----RGWKHPSGCI 341
+ + L R+F +RY D G V+ + S S P + R PSG +
Sbjct: 196 TYAPTTLAPARDFWTLRYTTTLDNGSLVVCERSLSGSGTGPNPTAAAQFVRAEMLPSGYL 375
Query: 342 IREMPHGGCLVTWVEHVEVE 361
IR GG ++ V+H+ +E
Sbjct: 376 IRPCDGGGSIIHIVDHLNLE 435
>TC90396 similar to PIR|H96719|H96719 homeobox gene 13 protein 11736-10437
[imported] - Arabidopsis thaliana, partial (41%)
Length = 997
Score = 41.6 bits (96), Expect = 0.001
Identities = 24/79 (30%), Positives = 41/79 (51%), Gaps = 11/79 (13%)
Frame = +1
Query: 9 EAERH-QLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKIRNENLKIK--- 64
E ER QLA +GL+P+Q+ WFQN+R K + + L+++ D ++ +N +K
Sbjct: 331 EPERKMQLAKALGLQPRQVAIWFQNRRARWKTKQLEKEYEVLKKQFDSLKADNNTLKAQN 510
Query: 65 -------EVLKAKICLDCG 76
+ LK + C + G
Sbjct: 511 NKLHAELQTLKKRDCFENG 567
>TC79846 similar to GP|8843728|dbj|BAA97276.1 homeodomain transcription
factor-like {Arabidopsis thaliana}, partial (42%)
Length = 679
Score = 41.6 bits (96), Expect = 0.001
Identities = 18/64 (28%), Positives = 38/64 (59%)
Frame = +2
Query: 5 HHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKIRNENLKIK 64
H + ++ +LA+E+GL+P+Q+ WFQN+R K++ E +L++ ++ E ++
Sbjct: 320 HKLESEKKDRLAMELGLDPRQVAVWFQNRRARWKNKKLEEEYFSLKKNHESTILEKCLLE 499
Query: 65 EVLK 68
L+
Sbjct: 500 TKLR 511
>TC85737 similar to GP|6091551|gb|AAF01764.2| homeodomain-leucine zipper
protein 56 {Glycine max}, partial (63%)
Length = 1622
Score = 40.8 bits (94), Expect = 0.002
Identities = 16/46 (34%), Positives = 28/46 (60%)
Frame = +2
Query: 12 RHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKIR 57
+ +LA+E+GL+P+Q+ WFQN+R K + G L+ D ++
Sbjct: 635 KEKLAIELGLQPRQVAVWFQNRRARWKTKQLERDYGVLKANYDALK 772
>BG584873 similar to GP|20161555|dbj putative homeodomain-leucine zipper
{Oryza sativa (japonica cultivar-group)}, partial (31%)
Length = 737
Score = 40.4 bits (93), Expect = 0.002
Identities = 28/117 (23%), Positives = 61/117 (51%), Gaps = 8/117 (6%)
Frame = +1
Query: 11 ERHQLAVEVGLEPKQIKFWFQNKR-------TLLKHQHERETNGTLRRENDKIRNENLKI 63
++ LA+++ L P+Q++ WFQN+R T ++ ++ + G+L +N +++ E +
Sbjct: 301 QKECLAMQLKLRPRQVEVWFQNRRARSKLKQTEMECEYLKRWFGSLTEQNRRLQRE---V 471
Query: 64 KEVLKAKICLDCGGPPFPMKDHQNFVQDLKQENAQLKQECEKM-SSLLASYMEKKIS 119
+E+ K+ GPP + H + + L + CE++ ++ A+ +KIS
Sbjct: 472 EELRAMKV-----GPPTVLSPHSS--EPLPASTLSMCPRCERVTTNTTAAVAAEKIS 621
>AW693872 similar to PIR|D84679|D846 probable homeodomain transcription
factor [imported] - Arabidopsis thaliana, partial (15%)
Length = 665
Score = 40.4 bits (93), Expect = 0.002
Identities = 24/109 (22%), Positives = 54/109 (49%), Gaps = 12/109 (11%)
Frame = +3
Query: 6 HPDEAERHQLAVEVGLEPKQIKFWFQNKRT-----LLKHQHERETNGTLRREN-DKIRNE 59
+P + ++H LA + GL Q+ WF N R +++ H E+ GT +N + +NE
Sbjct: 96 YPTDTDKHMLATQTGLSRNQVSNWFINARVRVWKPMVEEIHMLESKGTAEAQNQNNDKNE 275
Query: 60 NLKIKEVLKAKICLDCGGP------PFPMKDHQNFVQDLKQENAQLKQE 102
++ K CL+ G + + + ++ +L++++++L+ +
Sbjct: 276 ACRVGTHEKQFQCLEMGSSSGINNVEKVITNEEQWINNLQEKSSKLESD 422
>BM813592 similar to GP|20161555|db putative homeodomain-leucine zipper
{Oryza sativa (japonica cultivar-group)}, partial (42%)
Length = 683
Score = 40.4 bits (93), Expect = 0.002
Identities = 25/111 (22%), Positives = 57/111 (50%), Gaps = 7/111 (6%)
Frame = +2
Query: 5 HHPDEAERHQLAVEVGLEPKQIKFWFQNKR-------TLLKHQHERETNGTLRRENDKIR 57
H + ++ LA+++ L P+Q++ WFQN+R T ++ ++ + G+L +N +++
Sbjct: 242 HTLNPKQKECLAMQLKLRPRQVEVWFQNRRARSKLKQTEMECEYLKRWFGSLTEQNRRLQ 421
Query: 58 NENLKIKEVLKAKICLDCGGPPFPMKDHQNFVQDLKQENAQLKQECEKMSS 108
E ++E+ K+ GPP + H + + L + CE++++
Sbjct: 422 RE---VEELRAMKV-----GPPTVLSPHSS--EPLPASTLSMCPRCERVTT 544
>BG647110 homologue to PIR|T10695|T10 transcription factor HD-zip -
Arabidopsis thaliana, partial (16%)
Length = 844
Score = 40.0 bits (92), Expect = 0.003
Identities = 25/67 (37%), Positives = 33/67 (48%), Gaps = 4/67 (5%)
Frame = +1
Query: 3 ECHHPDEAERHQLAVEVGL----EPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKIRN 58
EC P R QL E + EPKQIK WFQN+R + +R+ G L+ N K+
Sbjct: 202 ECPKPTSLRRQQLIRECPILSHIEPKQIKVWFQNRRC---REKQRKEAGRLQAVNRKLTA 372
Query: 59 ENLKIKE 65
N + E
Sbjct: 373 MNKLLME 393
>TC79046 similar to PIR|T12634|T12634 homeotic protein - common sunflower,
partial (56%)
Length = 1136
Score = 38.9 bits (89), Expect = 0.006
Identities = 20/53 (37%), Positives = 31/53 (57%), Gaps = 1/53 (1%)
Frame = +2
Query: 9 EAERH-QLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKIRNEN 60
E ER QLA + L+P+Q+ WFQN+R K + + L+R+ D I+ +N
Sbjct: 470 EPERKMQLARALNLQPRQVAIWFQNRRARWKTKQLEKDYDVLKRQYDAIKLDN 628
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,946,764
Number of Sequences: 36976
Number of extensions: 326672
Number of successful extensions: 1447
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 1424
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1435
length of query: 657
length of database: 9,014,727
effective HSP length: 102
effective length of query: 555
effective length of database: 5,243,175
effective search space: 2909962125
effective search space used: 2909962125
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147010.5


