

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147007.6 - phase: 0 /pseudo
(1105 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
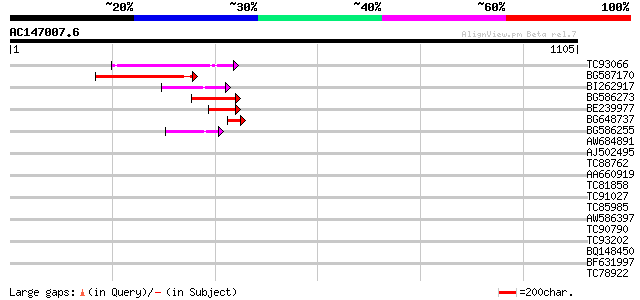
Score E
Sequences producing significant alignments: (bits) Value
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 160 3e-39
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 151 1e-36
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 96 1e-19
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 91 2e-18
BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryz... 79 1e-14
BG648737 weakly similar to GP|21433|emb|CA ORF3 {Solanum tuberos... 45 2e-04
BG586255 similar to GP|7682800|gb Hypothetical protein T15F17.l ... 44 5e-04
AW684891 41 0.003
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 38 0.025
TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Ar... 37 0.055
AA660919 similar to PIR|F84589|F84 probable protein kinase [impo... 37 0.055
TC81858 similar to GP|18252179|gb|AAL61922.1 unknown protein {Ar... 34 0.27
TC91027 similar to GP|14517502|gb|AAK62641.1 At2g38310/T19C21.20... 34 0.36
TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumb... 33 0.47
AW586397 33 0.61
TC90790 similar to GP|18086412|gb|AAL57663.1 At2g17250/T23A1.11 ... 28 0.80
TC93202 similar to PIR|T46039|T46039 hypothetical protein T16K5.... 32 1.4
BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.1... 31 2.3
BF631997 weakly similar to GP|18542925|gb Putative pol polyprote... 31 2.3
TC78922 similar to GP|4107276|emb|CAA67130.1 acetyl-CoA syntheta... 31 2.3
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 160 bits (404), Expect = 3e-39
Identities = 98/251 (39%), Positives = 143/251 (56%), Gaps = 4/251 (1%)
Frame = +1
Query: 199 GKHVRVSFPSRANKRSMSPFNIIHSDVWGPSRVSSTLGYKYYVTFIDDFSRCTWITLLKD 258
G +VSF S A R+ + IHSD+WGPS+V+S G +Y +T IDDF R W+ L+
Sbjct: 43 GNRKKVSF-STATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRY 219
Query: 259 RS*LFGAFQTFCS*IKTQFGKTIRILRSDNAK*YFSTSFNSFMASHGIIHQSSCPHTPQQ 318
++ F F+ + ++TQ GK ++ L +DN + S+ FN F +HGI + P PQQ
Sbjct: 220 KNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQ 399
Query: 319 NGVAERKHRHLVDTTRTLLINAHA--PFRFWGDAILIACYLITRMPSSVLGNEIPYSLLF 376
NGVAER R L++ R +L NA W +A AC+L+ R P S L ++P ++
Sbjct: 400 NGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPED-IW 576
Query: 377 PKDPLYVVPLRVFGSTCFAHDLSPDRDKLSARAVKCVFLGYSKTQKGYR--CYSPSAHRF 434
+ + LR+FG C A+ L D KL+ RA +C+FL Y+ KGYR C P + +
Sbjct: 577 SGNLVDYSNLRIFG--CPAYALVND-GKLAPRAGECIFLSYASESKGYRLWCSDPKSQKL 747
Query: 435 YVSKDVTFFED 445
+S+DVTF ED
Sbjct: 748 ILSRDVTFNED 780
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 151 bits (382), Expect = 1e-36
Identities = 80/200 (40%), Positives = 126/200 (63%)
Frame = -3
Query: 167 HCRLGHPSLDKLKVLVPHLSHLKSLDCESCQLGKHVRVSFPSRANKRSMSPFNIIHSDVW 226
H RLGHP L +++P + ++ +CE+C LGKH + FP R + + F++I++D+W
Sbjct: 578 HARLGHPHGRALNLMLPGVV-FENKNCEACILGKHCKNVFP-RTSTVYENCFDLIYTDLW 405
Query: 227 GPSRVSSTLGYKYYVTFIDDFSRCTWITLLKDRS*LFGAFQTFCS*IKTQFGKTIRILRS 286
+S +KY+VTFID+ S+ TW+TL+ + + AF+ F + + + I+ILRS
Sbjct: 404 TAPSLSRD-NHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKNFQAYVTNHYHAKIKILRS 228
Query: 287 DNAK*YFSTSFNSFMASHGIIHQSSCPHTPQQNGVAERKHRHLVDTTRTLLINAHAPFRF 346
DN Y S +F S + HGI+HQ+SCP+TPQQNGVA+RK++HL++ R+L+ A+
Sbjct: 227 DNGGEYTSYAFKSHLDHHGILHQTSCPYTPQQNGVAKRKNKHLMEVARSLMFQAN----- 63
Query: 347 WGDAILIACYLITRMPSSVL 366
+ ACYLI +P+ VL
Sbjct: 62 ----VSTACYLINWIPTKVL 15
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 95.5 bits (236), Expect = 1e-19
Identities = 53/134 (39%), Positives = 74/134 (54%)
Frame = +1
Query: 296 SFNSFMASHGIIHQSSCPHTPQQNGVAERKHRHLVDTTRTLLINAHAPFRFWGDAILIAC 355
SF F+ + S HTPQQNGVAER +R L++ TR +L A FW +A+ AC
Sbjct: 28 SF*HFVNKRVL*GNSXVAHTPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTAC 207
Query: 356 YLITRMPSSVLGNEIPYSLLFPKDPLYVVPLRVFGSTCFAHDLSPDRDKLSARAVKCVFL 415
Y+I R PS+V+ + P + K P+ L VFG + S +R KL ++ KC+FL
Sbjct: 208 YVINRSPSTVIDLKTPMEMWKGK-PVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFL 384
Query: 416 GYSKTQKGYRCYSP 429
GY+ KGY + P
Sbjct: 385 GYADNVKGYXLWDP 426
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 90.9 bits (224), Expect = 2e-18
Identities = 44/96 (45%), Positives = 62/96 (63%)
Frame = -2
Query: 354 ACYLITRMPSSVLGNEIPYSLLFPKDPLYVVPLRVFGSTCFAHDLSPDRDKLSARAVKCV 413
ACYLI R+P+ VL ++ P+ +L + P +RVFG C+ R+KL AR+ K +
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTY-MRVFGCLCYVLVPGELRNKLEARSRKAM 528
Query: 414 FLGYSKTQKGYRCYSPSAHRFYVSKDVTFFEDRPFF 449
F+GYS TQKGY+CY P A R VS+DV F E+R ++
Sbjct: 527 FIGYSTTQKGYKCYDPEARRVLVSRDVKFIEERGYY 420
>BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 514
Score = 78.6 bits (192), Expect = 1e-14
Identities = 34/63 (53%), Positives = 43/63 (67%)
Frame = -2
Query: 387 RVFGSTCFAHDLSPDRDKLSARAVKCVFLGYSKTQKGYRCYSPSAHRFYVSKDVTFFEDR 446
R+FG T F H S R K RA+KCVF+ YS TQKGYRCY P + +++VS+DVTF E
Sbjct: 456 RIFGCTSFVHIHSDGRSKFDHRALKCVFIRYSSTQKGYRCYHPPSRKYFVSRDVTFHEQE 277
Query: 447 PFF 449
+F
Sbjct: 276 SYF 268
>BG648737 weakly similar to GP|21433|emb|CA ORF3 {Solanum tuberosum}, partial
(10%)
Length = 804
Score = 44.7 bits (104), Expect = 2e-04
Identities = 18/35 (51%), Positives = 23/35 (65%)
Frame = +3
Query: 424 YRCYSPSAHRFYVSKDVTFFEDRPFFASPTTSGST 458
Y+CYSP +FY S DVTFFED+PF+ G +
Sbjct: 3 YKCYSPITKKFYNSMDVTFFEDQPFYTKIGIQGES 107
>BG586255 similar to GP|7682800|gb Hypothetical protein T15F17.l {Arabidopsis
thaliana}, partial (12%)
Length = 436
Score = 43.5 bits (101), Expect = 5e-04
Identities = 34/113 (30%), Positives = 51/113 (45%)
Frame = -1
Query: 305 GIIHQSSCPHTPQQNGVAERKHRHLVDTTRTLLINAHAPFRFWGDAILIACYLITRMPSS 364
G + +S H QNG+AE + + TR LL+ + P WG A+L A LI PSS
Sbjct: 415 G*VWNTSVVHVHTQNGLAESFIKRIQLITRPLLMRSKQPVSAWGHAVLHAAELIRIRPSS 236
Query: 365 VLGNEIPYSLLFPKDPLYVVPLRVFGSTCFAHDLSPDRDKLSARAVKCVFLGY 417
P LL P V ++ FG + P R K+ + +++G+
Sbjct: 235 E-HKYSPSQLLSGHVP-DVSHIKTFGYPVYVPIAPPHRTKMGPQRRMGIYVGF 83
>AW684891
Length = 539
Score = 40.8 bits (94), Expect = 0.003
Identities = 25/88 (28%), Positives = 43/88 (48%)
Frame = -3
Query: 236 GYKYYVTFIDDFSRCTWITLLKDRS*LFGAFQTFCS*IKTQFGKTIRILRSDNAK*YFST 295
G K+++ FI + + T + L K I QF K+I L S+ K Y
Sbjct: 348 GXKWFLAFIHNSTSIT*LFLKKHE-------------IWFQFVKSIERLCSNKGKVYVDQ 208
Query: 296 SFNSFMASHGIIHQSSCPHTPQQNGVAE 323
+ + ++ +G+ + C ++PQQNGV+E
Sbjct: 207 NLSQYLEENGVSQELKCVNSPQQNGVSE 124
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 37.7 bits (86), Expect = 0.025
Identities = 33/104 (31%), Positives = 48/104 (45%)
Frame = +3
Query: 956 IGQEMQVIDGLLLVIAFLWAEI*FHGRAKSRLLSPDRVQKPSIELWLILHVSLCG*NIYC 1015
IGQE+Q + F+ + +HG ++ L S QK +I L++ + CG* +
Sbjct: 6 IGQEIQKQEKARQGTRFI*EPVQYHGLRRNNLWSLFPQQKQNI*HQLVVLLKQCG*EEF* 185
Query: 1016 KNYIFVRLVKWNLYVIINQLYTSPPIQFFTRGQSILKLTVTFLE 1059
K I R + VI +Q IQFF G SIL + T E
Sbjct: 186 K*CIMSRTLLQRYIVITSQQLH*AKIQFFMDGPSILTSSFTRYE 317
>TC88762 similar to GP|22136928|gb|AAM91808.1 unknown protein {Arabidopsis
thaliana}, partial (3%)
Length = 805
Score = 36.6 bits (83), Expect = 0.055
Identities = 26/98 (26%), Positives = 38/98 (38%)
Frame = +3
Query: 449 FASPTTSGSTTSTTSTTDVTTSHVMPIPLFEPFVSTQNPPQSQGNPEFRQYGITYELRHV 508
F +PT+ S+ S +S TTS P P +T +PP S
Sbjct: 75 FNNPTSPSSSPSASSPVSTTTSPPASTPASSPVSTTTSPPAS------------------ 200
Query: 509 EAPETAPIDSNDSTPKTPATNSSDSGIVPVSSPAAVPP 546
P ++P+ + S P +S S P +SPA P
Sbjct: 201 -TPASSPVPTTTSPPAPTPASSPVSTNSPTASPAGSLP 311
>AA660919 similar to PIR|F84589|F84 probable protein kinase [imported] -
Arabidopsis thaliana, partial (24%)
Length = 579
Score = 36.6 bits (83), Expect = 0.055
Identities = 20/42 (47%), Positives = 26/42 (61%)
Frame = -2
Query: 149 LHSHPSTICGVSASPDIIHCRLGHPSLDKLKVLVPHLSHLKS 190
LHS T C +SA PD+ C L + +KLK++ PHL LKS
Sbjct: 152 LHSIYITSCLLSADPDLSKCTLPYHG-EKLKIISPHLMPLKS 30
>TC81858 similar to GP|18252179|gb|AAL61922.1 unknown protein {Arabidopsis
thaliana}, partial (9%)
Length = 748
Score = 34.3 bits (77), Expect = 0.27
Identities = 30/111 (27%), Positives = 41/111 (36%)
Frame = +1
Query: 436 VSKDVTFFEDRPFFASPTTSGSTTSTTSTTDVTTSHVMPIPLFEPFVSTQNPPQSQGNPE 495
+S +V FF A+ T G ST+ T SH P P +T PP S P
Sbjct: 76 MSSNVAFFFVFALLAT-TAFGEAPSTSPTAAPKASHAAPAPK-----ATATPPSSTTTPP 237
Query: 496 FRQYGITYELRHVEAPETAPIDSNDSTPKTPATNSSDSGIVPVSSPAAVPP 546
++ T+P S +P + + S PV SP PP
Sbjct: 238 -------------KSSATSPTSSPAPKVSSPPSPTPTSAEAPVESPTESPP 351
>TC91027 similar to GP|14517502|gb|AAK62641.1 At2g38310/T19C21.20
{Arabidopsis thaliana}, partial (55%)
Length = 682
Score = 33.9 bits (76), Expect = 0.36
Identities = 31/122 (25%), Positives = 49/122 (39%)
Frame = +2
Query: 428 SPSAHRFYVSKDVTFFEDRPFFASPTTSGSTTSTTSTTDVTTSHVMPIPLFEPFVSTQNP 487
+PS H+ +++ + + +P SPT S +TT T S TS P P S +
Sbjct: 260 TPSEHKLFLNNKICYQTQQP--PSPTPSPATTPTQSPP---TSVAPPSSNISPHPSPPSG 424
Query: 488 PQSQGNPEFRQYGITYELRHVEAPETAPIDSNDSTPKTPATNSSDSGIVPVSSPAAVPPE 547
P S + + P S+ + + T +S + VSSPA+ PP
Sbjct: 425 PSSVAST-------------IHKPTNT---SSKAAMSSSETATSALSVKSVSSPASPPPS 556
Query: 548 RP 549
P
Sbjct: 557 AP 562
>TC85985 similar to GP|5139695|dbj|BAA81686.1 expressed in cucumber
hypocotyls {Cucumis sativus}, partial (42%)
Length = 892
Score = 33.5 bits (75), Expect = 0.47
Identities = 33/108 (30%), Positives = 45/108 (41%), Gaps = 4/108 (3%)
Frame = +3
Query: 450 ASPTTSGSTTSTTSTTDVT--TSHVMPIPLFEPFVS--TQNPPQSQGNPEFRQYGITYEL 505
++PTTS T +T S V T P P+ P S +P + P
Sbjct: 132 SAPTTSPPTVTTPSAAPVAAPTKPKSPAPVASPKSSPPASSPTAATVTPAVSPAAPVPVA 311
Query: 506 RHVEAPETAPIDSNDSTPKTPATNSSDSGIVPVSSPAAVPPERPIDLP 553
+ A ++P+ + STP PA SS VPVSSP P P+ P
Sbjct: 312 KSPAA--SSPVVAPVSTPPKPAPVSSPPAPVPVSSP---PTPVPVSSP 440
>AW586397
Length = 646
Score = 33.1 bits (74), Expect = 0.61
Identities = 30/92 (32%), Positives = 44/92 (47%)
Frame = -1
Query: 26 STILQYACLTLLDHGF*ILVLLIMLLVIPLLYLICHLLRYLNITDANGYKAQVTGIGQAS 85
S L + ++ DH +* +VL I+ LV +YL Q T +GQAS
Sbjct: 289 SATLAHFGSSMWDHEY*TMVLFII*LV-----------KYLTTLPL----LQTTSVGQAS 155
Query: 86 PLPSLSLNYVLFISGSPFNLISISKLTQSLNC 117
PL S+ L+YV ++ PF +S L +NC
Sbjct: 154 PLSSVLLDYVYWV---PFLFDCVSVL--RVNC 74
>TC90790 similar to GP|18086412|gb|AAL57663.1 At2g17250/T23A1.11
{Arabidopsis thaliana}, partial (23%)
Length = 812
Score = 28.1 bits (61), Expect(2) = 0.80
Identities = 17/39 (43%), Positives = 23/39 (58%)
Frame = +2
Query: 518 SNDSTPKTPATNSSDSGIVPVSSPAAVPPERPIDLPIAF 556
SN + TP+++ S +P PAAVPP PI+ P AF
Sbjct: 188 SNRFSLFTPSSSLSSLNSLPPPPPAAVPPLTPIN-PNAF 301
Score = 23.1 bits (48), Expect(2) = 0.80
Identities = 8/21 (38%), Positives = 13/21 (61%)
Frame = +1
Query: 468 TTSHVMPIPLFEPFVSTQNPP 488
+ SH+ +PL F+S +PP
Sbjct: 115 SASHINNLPLLLTFISPSSPP 177
>TC93202 similar to PIR|T46039|T46039 hypothetical protein T16K5.80 -
Arabidopsis thaliana, partial (9%)
Length = 707
Score = 32.0 bits (71), Expect = 1.4
Identities = 17/35 (48%), Positives = 21/35 (59%)
Frame = -1
Query: 91 SLNYVLFISGSPFNLISISKLTQSLNCSITFSSDS 125
S+N +F S SP L+ S + SL CS TFSS S
Sbjct: 473 SVNVSMFTSSSPSMLLVSSASSSSLRCSSTFSSTS 369
>BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.18~similar to
unknown protein {Arabidopsis thaliana}, partial (9%)
Length = 666
Score = 31.2 bits (69), Expect = 2.3
Identities = 19/55 (34%), Positives = 31/55 (55%)
Frame = +1
Query: 492 GNPEFRQYGITYELRHVEAPETAPIDSNDSTPKTPATNSSDSGIVPVSSPAAVPP 546
GNP+FR R+ A +T I S+ +P + ++SS+ + P+SSP +PP
Sbjct: 85 GNPKFRN---PIS*RNTFACQTHAISSS*PSPNSHRSSSSNLTLSPLSSPK*LPP 240
>BF631997 weakly similar to GP|18542925|gb Putative pol polyprotein {Oryza
sativa}, partial (6%)
Length = 650
Score = 31.2 bits (69), Expect = 2.3
Identities = 16/47 (34%), Positives = 23/47 (48%)
Frame = -2
Query: 278 GKTIRILRSDNAK*YFSTSFNSFMASHGIIHQSSCPHTPQQNGVAER 324
G+ + + +N S FN I Q + +TPQ+NGVAER
Sbjct: 571 GRR*NVFKLNNGLEICSAEFNELCKEEHITRQYTVRNTPQKNGVAER 431
>TC78922 similar to GP|4107276|emb|CAA67130.1 acetyl-CoA synthetase {Solanum
tuberosum}, partial (84%)
Length = 1788
Score = 31.2 bits (69), Expect = 2.3
Identities = 27/98 (27%), Positives = 42/98 (42%), Gaps = 14/98 (14%)
Frame = +1
Query: 931 SKEHSEKDLFSLIEATQISLGIQM-----PIGQEMQVIDGLLLVIAFLWAEI*FHGR--- 982
+K+H L + I GI+M P M + +++ W ++*FH +
Sbjct: 1126 TKQHISSPLLATISVVMAVAGIKMDTIGLPEELTMSSMSVGIVLAQLKWNQL*FHTQNVP 1305
Query: 983 -----AKSRLLSPDRVQKPSIELWLILH-VSLCG*NIY 1014
A S L DRV P + LW++ H V CG +Y
Sbjct: 1306 KQPL*AWSTRLK-DRVYMPLLLLWMVFHTVKNCGRTLY 1416
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.342 0.151 0.501
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,748,081
Number of Sequences: 36976
Number of extensions: 661227
Number of successful extensions: 5705
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 3193
Number of HSP's successfully gapped in prelim test: 319
Number of HSP's that attempted gapping in prelim test: 2344
Number of HSP's gapped (non-prelim): 3766
length of query: 1105
length of database: 9,014,727
effective HSP length: 106
effective length of query: 999
effective length of database: 5,095,271
effective search space: 5090175729
effective search space used: 5090175729
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 64 (29.3 bits)
Medicago: description of AC147007.6


