

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147007.4 - phase: 0
(419 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
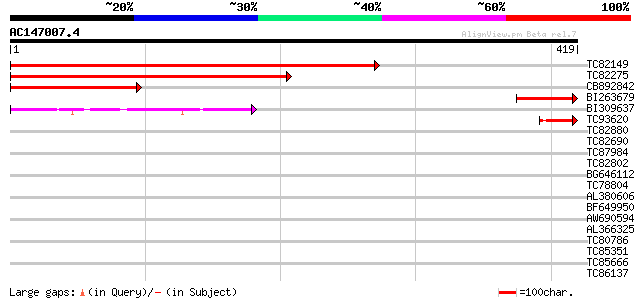
Score E
Sequences producing significant alignments: (bits) Value
TC82149 weakly similar to PIR|H96695|H96695 hypothetical protein... 384 e-107
TC82275 similar to PIR|H96695|H96695 hypothetical protein F5A8.1... 268 4e-72
CB892842 similar to PIR|H96695|H966 hypothetical protein F5A8.10... 126 1e-29
BI263679 97 9e-21
BI309637 similar to PIR|D86241|D86 protein T16B5.8 [imported] - ... 44 9e-05
TC93620 homologue to GP|12964610|dbj|BAB32671. pre-mRNA processi... 42 6e-04
TC82880 similar to GP|9858483|gb|AAG01054.1| resistance protein ... 38 0.008
TC82690 weakly similar to GP|9802755|gb|AAF99824.1| Unknown prot... 37 0.019
TC87984 weakly similar to GP|10177368|dbj|BAB10659. contains sim... 36 0.032
TC82802 similar to GP|9802755|gb|AAF99824.1| Unknown protein {Ar... 34 0.092
BG646112 weakly similar to GP|10177368|dbj contains similarity t... 34 0.12
TC78804 34 0.12
AL380606 33 0.20
BF649950 similar to GP|9802755|gb|A Unknown protein {Arabidopsis... 32 0.46
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 32 0.60
AL366325 31 1.0
TC80786 similar to GP|20339360|gb|AAM19353.1 TIR-similar-domain-... 31 1.0
TC85351 30 1.3
TC85666 homologue to SP|P10933|FENR_PEA Ferredoxin--NADP reducta... 30 2.3
TC86137 weakly similar to PIR|A96673|A96673 probable cytochrome ... 29 3.0
>TC82149 weakly similar to PIR|H96695|H96695 hypothetical protein F5A8.10
[imported] - Arabidopsis thaliana, partial (63%)
Length = 1225
Score = 384 bits (987), Expect = e-107
Identities = 193/273 (70%), Positives = 225/273 (81%)
Frame = +3
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
M++LPVEV+G ILS LGSARDV+IASLTCKKWR+AW+ HL TL F T DWP+YRE+ SS
Sbjct: 402 MDNLPVEVIGNILSLLGSARDVVIASLTCKKWRQAWRYHLHTLEFITFDWPVYRELSSST 581
Query: 61 LEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRYNVRTSPNFNII 120
LE++IT T+FQTK L+CLTI MDD HEFS VIAW MYTR+ LR L + V T P FNII
Sbjct: 582 LEIIITQTIFQTKALRCLTIKMDDVHEFSAASVIAWFMYTREDLRRLHFYVNTPPMFNII 761
Query: 121 EKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSISALDLSLLLSACPKLETLS 180
EKC+R+ LEVL L N I+ VEP Y KFPCLKSLSLSFVSISALDLSLLLS CP+LETL+
Sbjct: 762 EKCARQKLEVLVLGQNFITRVEPSYLKFPCLKSLSLSFVSISALDLSLLLSVCPRLETLA 941
Query: 181 IVCPEIAMSDSEASIELSSSSLKDFFVESYSFDKLILVADMLENLHLKDCSFDAFELINK 240
+V PEIAMSDS+AS+ELSSSSLK+F VES+ DK +L AD+LE LHLKDC+F+ FE I K
Sbjct: 942 LVSPEIAMSDSQASMELSSSSLKEFSVESFGLDKFVLEADLLECLHLKDCTFEVFEFIGK 1121
Query: 241 GTLKVLKLDDVSVIHLDIGDNTENLEIVDVCNF 273
LKVLK+DDVSVIHLDIG+N +NLE VD+ NF
Sbjct: 1122EGLKVLKVDDVSVIHLDIGENADNLEYVDISNF 1220
>TC82275 similar to PIR|H96695|H96695 hypothetical protein F5A8.10
[imported] - Arabidopsis thaliana, partial (49%)
Length = 751
Score = 268 bits (684), Expect = 4e-72
Identities = 133/208 (63%), Positives = 167/208 (79%)
Frame = +1
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
M+ LPVEV+G ILSHL SARDV+IAS TCKKWR A HL TLSF+++DW +YR++ ++
Sbjct: 127 MDQLPVEVIGNILSHLKSARDVVIASATCKKWRTACCKHLHTLSFSSNDWSVYRDLSTTR 306
Query: 61 LEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRYNVRTSPNFNII 120
LE+LIT T+FQT GLQ L+I M+D EFS + VIAWLMYTR++LR+L YNV+T PN NI+
Sbjct: 307 LEILITQTIFQTSGLQSLSILMEDVDEFSASAVIAWLMYTRETLRQLFYNVKTMPNVNIL 486
Query: 121 EKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSISALDLSLLLSACPKLETLS 180
E C R LE+L L N I GVEP Y +FPCLKSLSLS VSISALDL+LL+SACP++E L
Sbjct: 487 EICGRHKLEILDLEHNSIVGVEPNYQRFPCLKSLSLSCVSISALDLNLLVSACPRIEALE 666
Query: 181 IVCPEIAMSDSEASIELSSSSLKDFFVE 208
+V PEIAMSD+ ++ELSSS+LK+ +VE
Sbjct: 667 LVNPEIAMSDAMVTVELSSSTLKNVYVE 750
>CB892842 similar to PIR|H96695|H966 hypothetical protein F5A8.10 [imported]
- Arabidopsis thaliana, partial (23%)
Length = 846
Score = 126 bits (317), Expect = 1e-29
Identities = 61/97 (62%), Positives = 77/97 (78%)
Frame = +3
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYREICSSH 60
M+ LPVEV+G ILSHL SARDV+IAS TCKKWR A HL TLSF+++DW +YR++ ++
Sbjct: 555 MDQLPVEVIGNILSHLKSARDVVIASATCKKWRTACCKHLHTLSFSSNDWSVYRDLSTTR 734
Query: 61 LEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWL 97
LE+LIT T+FQT GLQ L+I M+D EFS + VIAWL
Sbjct: 735 LEILITQTIFQTSGLQSLSILMEDVDEFSASAVIAWL 845
>BI263679
Length = 482
Score = 97.4 bits (241), Expect = 9e-21
Identities = 45/45 (100%), Positives = 45/45 (100%)
Frame = +3
Query: 375 KKMVIYGYVAEIKTHEECQTFTKFTEFMIQLGRKYSHIKFEFEYE 419
KKMVIYGYVAEIKTHEECQTFTKFTEFMIQLGRKYSHIKFEFEYE
Sbjct: 12 KKMVIYGYVAEIKTHEECQTFTKFTEFMIQLGRKYSHIKFEFEYE 146
>BI309637 similar to PIR|D86241|D86 protein T16B5.8 [imported] - Arabidopsis
thaliana, partial (28%)
Length = 714
Score = 44.3 bits (103), Expect = 9e-05
Identities = 45/192 (23%), Positives = 85/192 (43%), Gaps = 10/192 (5%)
Frame = +2
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSF--NTSDWPLYREICS 58
+E LP ++ ILSH+ + RDV + K+W+ + +++L F N D +RE
Sbjct: 62 IESLPDVILQYILSHVSNGRDVAYCNCVSKRWKNS-MACIRSLYFTRNAFDNAPHRE--- 229
Query: 59 SHLEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYTRDSLRELRYNVRTSPNFN 118
+ ++++ + + L+ L ++ FSV + +WL SL L + +
Sbjct: 230 -NSDIIVKRMVSAVERLEELVVYC----PFSVYGLASWLSLAGPSLSHLELRMDNLGDNE 394
Query: 119 IIEKCSRK--------TLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSISALDLSLLL 170
II + K +E L L I + PK+ F L+ L + + ++ ++
Sbjct: 395 IIHESPSKLDCIGAAVNVETLKLWGVLIKLI-PKWETFHNLRILEVVGARVEDAAVNAMI 571
Query: 171 SACPKLETLSIV 182
ACP L L ++
Sbjct: 572 QACPNLTRLLLL 607
>TC93620 homologue to GP|12964610|dbj|BAB32671. pre-mRNA processing 8
protein {Mus musculus}, partial (1%)
Length = 446
Score = 41.6 bits (96), Expect = 6e-04
Identities = 20/28 (71%), Positives = 22/28 (78%)
Frame = +1
Query: 392 CQTFTKFTEFMIQLGRKYSHIKFEFEYE 419
CQ KFTEF+IQLGR+Y IK EFEYE
Sbjct: 157 CQA--KFTEFIIQLGREYMDIKIEFEYE 234
>TC82880 similar to GP|9858483|gb|AAG01054.1| resistance protein MG55
{Glycine max}, partial (46%)
Length = 1238
Score = 37.7 bits (86), Expect = 0.008
Identities = 40/169 (23%), Positives = 71/169 (41%), Gaps = 8/169 (4%)
Frame = +2
Query: 136 NPISGVEPKYHKFPCLKSLSLSFVSISALDLSLLLSACPKLETLSIVCPEIAMSDSEASI 195
N ++ + P L SL + +++ + S +LS+C KL ++S+ C DSE +
Sbjct: 749 NSVAHISPFGGMSKSLASLDIESNNLALVYQSQILSSCSKLRSVSVQC------DSEIQL 910
Query: 196 ELSSSSLKDFFVESYSFDKLILVADMLENLHLKDCSFDAFELINKGTLKVL------KLD 249
+ + F + Y D + + H+ D S + LI G ++ L
Sbjct: 911 K---QEFRRFLDDLY--DAGLTELGISHASHISDHSLRSL-LIGMGNCHIVINILGKSLS 1072
Query: 250 DV--SVIHLDIGDNTENLEIVDVCNFIFMWQNFYNMISKASKLKKLRLW 296
V HL EN+E+ +C F F W F N + S + ++ +W
Sbjct: 1073QVPSQSSHLIFYHY*ENMELCSICMFFFKW*RFIN-VHFYSSIYQILIW 1216
>TC82690 weakly similar to GP|9802755|gb|AAF99824.1| Unknown protein
{Arabidopsis thaliana}, partial (12%)
Length = 762
Score = 36.6 bits (83), Expect = 0.019
Identities = 49/190 (25%), Positives = 77/190 (39%), Gaps = 8/190 (4%)
Frame = +1
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNT---SDWPLYREIC 57
+ LP ++ ILS L ++ + S+ KWR W T L L F++ SD +
Sbjct: 121 ISSLPGHIIDQILSIL-PIKEAVRTSILSTKWRYKWAT-LPNLVFDSQCISDTSEDLLVI 294
Query: 58 SSHLEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMY-TRDSLRELRYNVRTSPN 116
S L +I L G + VT + W ++ TR ++E +
Sbjct: 295 KSKLSRIIDHVLLLHSG-PIKKFKLSHRELIGVTDIDRWTLHLTRRPVKEFVLEIWKGQR 471
Query: 117 FNI---IEKCSR-KTLEVLTLASNPISGVEPKYHKFPCLKSLSLSFVSISALDLSLLLSA 172
+ I + C LE+ P S + F LKSL L V++S L+S
Sbjct: 472 YKIPSCLFSCQGLHHLELFNCWLIPPS----TFQGFRNLKSLDLQHVTLSQDAFENLIST 639
Query: 173 CPKLETLSIV 182
CP LE L+++
Sbjct: 640 CPLLERLTLM 669
>TC87984 weakly similar to GP|10177368|dbj|BAB10659. contains similarity to
heat shock transcription factor HSF30~gene_id:K16L22.13,
partial (5%)
Length = 696
Score = 35.8 bits (81), Expect = 0.032
Identities = 47/195 (24%), Positives = 77/195 (39%), Gaps = 14/195 (7%)
Frame = +3
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWPLYRE----- 55
+ LP ++ ILS L + V L +KWR W+ +L+ L F S P E
Sbjct: 111 INQLPNSLLHHILSFLPTKTCVQTTPLVSRKWRNLWK-NLEALDFCDSSSPQIYEFDNDE 287
Query: 56 ---ICSSHLEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVIAWLMYT-RDSLRELRYNV 111
I S + ++T + CL+ + F + W+ T +L E +
Sbjct: 288 QFLIFSVFVNTVLTLRKSRVVRKFCLSCYHVQLDPFYNHSIDTWINATIGPNLEEFHLTL 467
Query: 112 RTSPNFNIIEKCSRKTLEVLTLASNPISGVEPKYHKFPCLKSLS-LSFVSISALDLSL-- 168
T+ FN + +++L+ N ++ + + CL SL L + + LDL
Sbjct: 468 LTAAGFNRVPLSLFSCPNLVSLSFNDYIILQLQDNSKICLPSLKLLQLLDMYNLDLKFCE 647
Query: 169 --LLSACPKLETLSI 181
A P LE L I
Sbjct: 648 MPFSLAVPVLENLEI 692
>TC82802 similar to GP|9802755|gb|AAF99824.1| Unknown protein {Arabidopsis
thaliana}, partial (50%)
Length = 864
Score = 34.3 bits (77), Expect = 0.092
Identities = 32/111 (28%), Positives = 51/111 (45%), Gaps = 4/111 (3%)
Frame = +3
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSDWP--LYREICS 58
+ DLP ++ IL L RD + S+ +KWR W T Q L F+ +P RE+
Sbjct: 138 ISDLPQSIIETILIQL-PIRDAVRTSILSRKWRYKWSTITQ-LVFDEKCYPNSTDREVVQ 311
Query: 59 SHLEMLITCTLFQTKGLQCLTIFMDDEHEFSVTPVI-AWLMY-TRDSLREL 107
+ IT LF +G + F P I W+++ +R+ L++L
Sbjct: 312 KSVVEFITRLLFLHQG--PIHKFQIVNTSLQTCPAINQWILFLSRNDLKDL 458
>BG646112 weakly similar to GP|10177368|dbj contains similarity to heat shock
transcription factor HSF30~gene_id:K16L22.13, partial
(5%)
Length = 742
Score = 33.9 bits (76), Expect = 0.12
Identities = 50/205 (24%), Positives = 83/205 (40%), Gaps = 26/205 (12%)
Frame = +2
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSF------NTSDWPLYR 54
M LP ++ ILS L + V+ SL ++WR W+ HL F N + +R
Sbjct: 20 MSSLPDSLLCHILSFLPTKTSVMTTSLVSRRWRHLWE-HLNVFVFDDKSNCNCRNPKKFR 196
Query: 55 EICSSHLEMLITCTLFQTKGLQ--CLTIFMDDEHEFSVTPVIAWLMYT-RDSLRELRYNV 111
+ +L +L +++ ++ LT D + F V W+ L +L N+
Sbjct: 197 KFAFFVSSVL---SLRKSRHIRKFHLTCCTSDVYSFPGECVYMWVRAAIGPHLEDLSLNI 367
Query: 112 RTSPNFNIIEKCSRKTLEVLTLASNPISGV-----EPKYHKFPCLKSLSLSFVSIS-ALD 165
T+ + + + L L S + G+ +P FP LK L + F + +D
Sbjct: 368 -TNCHGDDMVYLPPSLLNCTNLVSLSLFGLIHLKFQPSAIHFPSLKMLKVEFSILEHNID 544
Query: 166 LS-----------LLLSACPKLETL 179
+ + LS CP LETL
Sbjct: 545 IEDFGKHLTDSILVFLSGCPVLETL 619
>TC78804
Length = 1318
Score = 33.9 bits (76), Expect = 0.12
Identities = 53/214 (24%), Positives = 87/214 (39%), Gaps = 14/214 (6%)
Frame = +3
Query: 26 SLTCKKWREAWQTHLQTLSFNT---SDWPLYREICSSHLEMLITCTLFQTKGLQCLTIFM 82
S+ WR W T L L F+ S+ + S L +I L G + F
Sbjct: 21 SILSTNWRYKWTT-LPNLVFDNECLSETSKDLLVIKSKLSRIIDHVLLLRSGP--IKKFK 191
Query: 83 DDEHEFSVTPVIAWLMY-TRDSLRELRYNVRTSPNFNI-----IEKC-SRKTLEVLTLAS 135
VT + W +Y TR ++E + +P+ + C S LE+L
Sbjct: 192 LSRDHIDVTDIDRWTLYLTRWQVKEFVLEIWKNPDQRYKIPSWLFSCQSLHHLELLECWL 371
Query: 136 NPISGVEPKYHKFPCLKSLSLSFVSISALDLSLLLSACPKLETLSIVCPEIAMSDSEASI 195
P S + F LKSL L +V+++ + L+S+C LE L ++ D ++
Sbjct: 372 IPPS----TFQGFKNLKSLDLQYVTLTKDAVENLISSCLMLERLILM-----ECDGFKNL 524
Query: 196 ELSSSSLKDFFV----ESYSFDKLILVADMLENL 225
+ + +L+ F + E SF +AD+ NL
Sbjct: 525 NIHAPNLQYFLMEGKFEDISFKNTSQLADVFINL 626
>AL380606
Length = 437
Score = 33.1 bits (74), Expect = 0.20
Identities = 17/36 (47%), Positives = 23/36 (63%), Gaps = 1/36 (2%)
Frame = +1
Query: 147 KFPCLKSLSLSFVSISAL-DLSLLLSACPKLETLSI 181
+FP LK+LSL + S + D LLL+ CP LE L +
Sbjct: 274 EFPSLKTLSLRLIKFSEVRDFMLLLAGCPILEDLHV 381
>BF649950 similar to GP|9802755|gb|A Unknown protein {Arabidopsis thaliana},
partial (7%)
Length = 581
Score = 32.0 bits (71), Expect = 0.46
Identities = 30/97 (30%), Positives = 46/97 (46%), Gaps = 2/97 (2%)
Frame = +2
Query: 87 EFSVTPVIAWLMY-TRDSLRELRYNVRTSPNFNIIEKCSRKTLEVLTLASNPISGVEPKY 145
E V+ V W++Y +R S++EL +V T + I C + L + P
Sbjct: 206 ESLVSDVDQWILYLSRKSIKELVLDVCTEELYKI-PWCLFSCQSLHHLKLHFCCLKPPTL 382
Query: 146 HK-FPCLKSLSLSFVSISALDLSLLLSACPKLETLSI 181
K F LKSL L+ V+++ L+S CP LE L +
Sbjct: 383 FKGFRSLKSLDLNHVAVAQDAFENLISGCPLLEKLKL 493
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 31.6 bits (70), Expect = 0.60
Identities = 18/46 (39%), Positives = 24/46 (52%)
Frame = +3
Query: 4 LPVEVVGIILSHLGSARDVLIASLTCKKWREAWQTHLQTLSFNTSD 49
LP ++ ILS L + V SL +KWR W+ LQ +F SD
Sbjct: 270 LPDPLLLHILSFLPTRTSVATMSLASRKWRNLWE-QLQVFNFKDSD 404
>AL366325
Length = 195
Score = 30.8 bits (68), Expect = 1.0
Identities = 17/32 (53%), Positives = 19/32 (59%)
Frame = +3
Query: 1 MEDLPVEVVGIILSHLGSARDVLIASLTCKKW 32
ME LP EVV ILS L S R++L CK W
Sbjct: 36 MEHLPQEVVSNILSRLPS-RELLKCKFVCKSW 128
>TC80786 similar to GP|20339360|gb|AAM19353.1 TIR-similar-domain-containing
protein TSDC {Pisum sativum}, partial (20%)
Length = 831
Score = 30.8 bits (68), Expect = 1.0
Identities = 15/52 (28%), Positives = 21/52 (39%)
Frame = +1
Query: 34 EAWQTHLQTLSFNTSDWPLYREICSSHLEMLITCTLFQTKGLQCLTIFMDDE 85
E W+ L L +T+ W ++ T +LFQ FMDDE
Sbjct: 277 EEWEKSLHNLRHSTAQWQMFLSFRGEDTRYSFTGSLFQALSQGGFKTFMDDE 432
>TC85351
Length = 720
Score = 30.4 bits (67), Expect = 1.3
Identities = 14/25 (56%), Positives = 15/25 (60%)
Frame = -3
Query: 351 LGWTTISDLFSVWVAGLLEGCPNLK 375
LGW + F VWV G LEG PN K
Sbjct: 637 LGWQMMEH-FPVWVVGFLEGWPNCK 566
>TC85666 homologue to SP|P10933|FENR_PEA Ferredoxin--NADP reductase leaf
isozyme chloroplast precursor (EC 1.18.1.2) (FNR).
[Garden pea], partial (96%)
Length = 1389
Score = 29.6 bits (65), Expect = 2.3
Identities = 16/49 (32%), Positives = 27/49 (54%)
Frame = +3
Query: 284 ISKASKLKKLRLWSVVFDDEDEVVDIETISVCFPRLTHLSLSYDLKDGV 332
I K K KLRL+S+ + D +T+S+C RL + + + ++ GV
Sbjct: 477 IDKNGKPHKLRLYSIASSALGDFGDSKTVSLCVKRLVYTNDAGEVVKGV 623
>TC86137 weakly similar to PIR|A96673|A96673 probable cytochrome P450
F13O11.25 [imported] - Arabidopsis thaliana, partial
(69%)
Length = 1695
Score = 29.3 bits (64), Expect = 3.0
Identities = 25/87 (28%), Positives = 38/87 (42%), Gaps = 4/87 (4%)
Frame = +1
Query: 98 MYTRDSLRELRYNVRTSPNFNIIEKCSRKTLEVLTLASNPISGVEPKYH----KFPCLKS 153
+YT S + + SPN N+ K S + + L+S IS ++ + +P + S
Sbjct: 169 LYTSPSSQTSNCSKNQSPNLNLFSKLS---MPNMVLSSLSISVLDLPFSLTTILWPTMFS 339
Query: 154 LSLSFVSISALDLSLLLSACPKLETLS 180
S+ AL L LL CP T S
Sbjct: 340 FKTHPSSLIALQLFLLPKCCPATNTTS 420
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.138 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,262,860
Number of Sequences: 36976
Number of extensions: 244969
Number of successful extensions: 1287
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 1263
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1281
length of query: 419
length of database: 9,014,727
effective HSP length: 99
effective length of query: 320
effective length of database: 5,354,103
effective search space: 1713312960
effective search space used: 1713312960
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC147007.4


