

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147007.11 - phase: 0
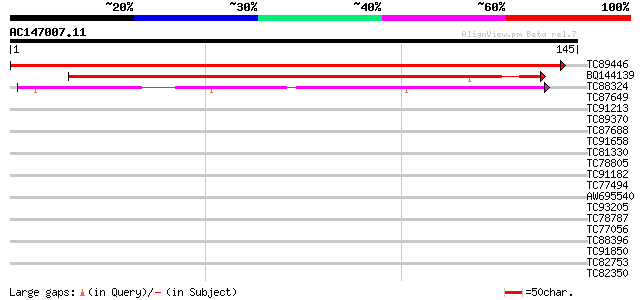
(145 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89446 similar to GP|21592451|gb|AAM64402.1 unknown {Arabidopsi... 276 2e-75
BQ144139 145 4e-36
TC88324 similar to PIR|F86238|F86238 protein T10O24.11 [imported... 66 4e-12
TC87649 similar to SP|Q9SZR1|ACAA_ARATH Potential calcium-transp... 31 0.20
TC91213 weakly similar to PIR|G71405|G71405 hypothetical protein... 30 0.26
TC89370 weakly similar to PIR|T01856|T01856 hypothetical protein... 29 0.58
TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~un... 29 0.58
TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome... 29 0.76
TC81330 homologue to GP|20466796|gb|AAM20715.1 unknown protein {... 28 0.99
TC78805 similar to PIR|T05236|T05236 hypothetical protein F18A5.... 28 1.7
TC91182 homologue to SP|Q9XQU4|SECY_PEA Preprotein translocase s... 28 1.7
TC77494 similar to SP|P21421|RPOB_PLAFA DNA-directed RNA polymer... 28 1.7
AW695540 similar to GP|22597166|gb| RING-H2 finger protein {Glyc... 27 2.2
TC93205 similar to PIR|A86309|A86309 hypothetical protein AAD500... 27 2.2
TC78787 similar to GP|13569996|gb|AAK31280.1 putative Myb-relate... 27 2.2
TC77056 similar to GP|15982817|gb|AAL09756.1 At2g20760/F5H14.27 ... 27 2.2
TC88396 weakly similar to GP|18616497|emb|CAD22847. unnamed prot... 27 2.2
TC91850 similar to GP|22597166|gb|AAN03470.1 RING-H2 finger prot... 27 2.2
TC82753 similar to GP|9758920|dbj|BAB09457.1 gb|AAD25781.1~gene_... 27 2.9
TC82350 similar to GP|9837385|gb|AAG00554.1| retinitis pigmentos... 27 2.9
>TC89446 similar to GP|21592451|gb|AAM64402.1 unknown {Arabidopsis
thaliana}, partial (54%)
Length = 747
Score = 276 bits (706), Expect = 2e-75
Identities = 140/142 (98%), Positives = 142/142 (99%)
Frame = +3
Query: 1 MQTSLFRRLAPLLAARFQRNQNIISSFSSSSSSSVLHHATSSPSSPDVEPVHITENCVRK 60
MQTSLFRRLAPLLAARFQRNQNIISSFSSSSSSSVLHHATSSPSSPDVEPVHITENCVRK
Sbjct: 45 MQTSLFRRLAPLLAARFQRNQNIISSFSSSSSSSVLHHATSSPSSPDVEPVHITENCVRK 224
Query: 61 MKELDANESSSGGKMLRLSVETGGCSGFQYAFNLDDRFNSDDRVFEKEGIKLVVDNISYD 120
MKELDANESSSGGKMLRLSVETGGCSGFQYAFNLDDRFNSDDRVFEKEGIKLVVDNISYD
Sbjct: 225 MKELDANESSSGGKMLRLSVETGGCSGFQYAFNLDDRFNSDDRVFEKEGIKLVVDNISYD 404
Query: 121 FVKGATVDYVEELIRSAFIVSQ 142
FVKGATVDYVEELIRSAFIV++
Sbjct: 405 FVKGATVDYVEELIRSAFIVTE 470
>BQ144139
Length = 685
Score = 145 bits (367), Expect = 4e-36
Identities = 80/123 (65%), Positives = 91/123 (73%), Gaps = 1/123 (0%)
Frame = +3
Query: 16 RFQRNQNIISSFSSSSSSSVLHHATSSPSSPDVEPVHITENCVRKMKELDANESSSGGKM 75
R QR QNII S SSSSSS+L+HA+ SP PDVEP HIT+NC R MKEL ANE SSGGKM
Sbjct: 96 RLQRTQNIIISI*SSSSSSLLYHASYSPDCPDVEPAHITQNCARIMKELYANE*SSGGKM 275
Query: 76 LRLSVETGGCSGFQYAFNLDDRFNSDDRVFEKEGIKLVVDN-ISYDFVKGATVDYVEELI 134
L V TGGCSGFQ A+N DDR+NSDD++F+KEGIKLVVDN + KG+T E L
Sbjct: 276 LL*IV*TGGCSGFQDAYNSDDRYNSDDKIFDKEGIKLVVDNMVIQPLAKGST----ERLC 443
Query: 135 RSA 137
R A
Sbjct: 444 RGA 452
>TC88324 similar to PIR|F86238|F86238 protein T10O24.11 [imported] -
Arabidopsis thaliana, partial (95%)
Length = 815
Score = 66.2 bits (160), Expect = 4e-12
Identities = 48/140 (34%), Positives = 71/140 (50%), Gaps = 4/140 (2%)
Frame = +1
Query: 3 TSL-FRRLAPLLAARFQRNQNIISSFSSSSSSSVLHHATSSPSSPDVEP-VHITENCVRK 60
TSL FR P+L RF I S S S+S++ P+S V P + +T+N +
Sbjct: 130 TSLSFRSSTPILNRRFSPKPLSIRSLSVSASAA--------PASGAVAPAISLTDNALNH 285
Query: 61 MKELDANESSSGGKMLRLSVETGGCSGFQYAFNLDDRFNS--DDRVFEKEGIKLVVDNIS 118
+ ++ + S LR+ V+ GGCSG Y + +DR N+ DD + E +G +V D S
Sbjct: 286 LNKMRSERSQD--LCLRIGVKQGGCSGMSYTMDFEDRANARPDDSIIEYKGFVIVCDPKS 459
Query: 119 YDFVKGATVDYVEELIRSAF 138
FV G +DY + LI F
Sbjct: 460 LLFVFGMQLDYSDALIGGGF 519
>TC87649 similar to SP|Q9SZR1|ACAA_ARATH Potential calcium-transporting ATPase
10 plasma membrane-type (EC 3.6.3.8), partial (43%)
Length = 2159
Score = 30.8 bits (68), Expect = 0.20
Identities = 23/81 (28%), Positives = 42/81 (51%), Gaps = 4/81 (4%)
Frame = -3
Query: 67 NESSSGGKMLRLSVETGGCSGFQYAFNLDD-RFNSDDRVFEKEGIKLVVDNISYD---FV 122
NES+S L ++ GGCS FN D+ +++S+ +E VV + S++ F+
Sbjct: 1434 NESNSDRNKPLLPIKPGGCSELPKKFNYDNLKYDSESHYPHEE----VVSSNSFENVVFI 1267
Query: 123 KGATVDYVEELIRSAFIVSQA 143
A + ++E+L + I Q+
Sbjct: 1266 WLARIKFIEDLTDNKCIEDQS 1204
>TC91213 weakly similar to PIR|G71405|G71405 hypothetical protein -
Arabidopsis thaliana, partial (24%)
Length = 836
Score = 30.4 bits (67), Expect = 0.26
Identities = 15/31 (48%), Positives = 21/31 (67%)
Frame = +1
Query: 26 SFSSSSSSSVLHHATSSPSSPDVEPVHITEN 56
S SS +SSS +HHA +S D+E VH+ +N
Sbjct: 229 SVSSMNSSSNIHHAQGE-TSRDIEKVHVDDN 318
>TC89370 weakly similar to PIR|T01856|T01856 hypothetical protein F9D12.19 -
Arabidopsis thaliana, partial (15%)
Length = 710
Score = 29.3 bits (64), Expect = 0.58
Identities = 20/47 (42%), Positives = 27/47 (56%), Gaps = 3/47 (6%)
Frame = +1
Query: 24 ISSFSSSSSSSVLHH---ATSSPSSPDVEPVHITENCVRKMKELDAN 67
+SS SSSSSSS +HH + ++ SS +P + T N KE D N
Sbjct: 154 LSSSSSSSSSSSIHHRLVSVAASSSTQTQPRNNTNN----KKEEDIN 282
>TC87688 similar to GP|10177535|dbj|BAB10930. gene_id:K1F13.21~unknown
protein {Arabidopsis thaliana}, partial (46%)
Length = 2077
Score = 29.3 bits (64), Expect = 0.58
Identities = 23/73 (31%), Positives = 35/73 (47%), Gaps = 3/73 (4%)
Frame = -2
Query: 1 MQTSLFRRLA---PLLAARFQRNQNIISSFSSSSSSSVLHHATSSPSSPDVEPVHITENC 57
+ +S+F+ L+ P L F + + SS SSSSSS ++S SSP + P +
Sbjct: 603 VNSSIFKNLSSIPPPLTFSFSSSSSSSSSLPSSSSSS----SSSKSSSPSLPPFAFFFST 436
Query: 58 VRKMKELDANESS 70
K +N SS
Sbjct: 435 PSKSSSSSSNSSS 397
>TC91658 similar to GP|14329812|emb|CAC40753. putative nucleosome assembly
protein 1 {Atropa belladonna}, partial (46%)
Length = 583
Score = 28.9 bits (63), Expect = 0.76
Identities = 19/38 (50%), Positives = 22/38 (57%)
Frame = -2
Query: 9 LAPLLAARFQRNQNIISSFSSSSSSSVLHHATSSPSSP 46
L PLL F + SS SSSSSSS ++SS SSP
Sbjct: 216 LLPLLLLLFPFPSSSSSSSSSSSSSSSSSSSSSSSSSP 103
>TC81330 homologue to GP|20466796|gb|AAM20715.1 unknown protein {Arabidopsis
thaliana}, partial (11%)
Length = 698
Score = 28.5 bits (62), Expect = 0.99
Identities = 14/35 (40%), Positives = 20/35 (57%)
Frame = +1
Query: 12 LLAARFQRNQNIISSFSSSSSSSVLHHATSSPSSP 46
LLA R +++Q++ S S+SV HH P SP
Sbjct: 577 LLAPRIKKSQHLSKIVLPSKSASVTHHILYLPVSP 681
>TC78805 similar to PIR|T05236|T05236 hypothetical protein F18A5.60 -
Arabidopsis thaliana, partial (21%)
Length = 717
Score = 27.7 bits (60), Expect = 1.7
Identities = 20/76 (26%), Positives = 31/76 (40%)
Frame = +1
Query: 30 SSSSSVLHHATSSPSSPDVEPVHITENCVRKMKELDANESSSGGKMLRLSVETGGCSGFQ 89
S S+++ +S P P I +N K D S+GG + R+ G S +
Sbjct: 106 SHFSNLIFPPINSIPLPLTAPFPIVKNTAGSAKNNDGFARSNGGFVRRIDGTVNGLSFSE 285
Query: 90 YAFNLDDRFNSDDRVF 105
NL+ +FN F
Sbjct: 286 RFLNLNFKFNLSSTEF 333
>TC91182 homologue to SP|Q9XQU4|SECY_PEA Preprotein translocase secY subunit
chloroplast precursor (CpSecY). [Garden pea] {Pisum
sativum}, partial (51%)
Length = 999
Score = 27.7 bits (60), Expect = 1.7
Identities = 20/53 (37%), Positives = 26/53 (48%), Gaps = 1/53 (1%)
Frame = +2
Query: 20 NQNIISSFSSSSSSSVLHHATSSPSSPDVEPVHITENC-VRKMKELDANESSS 71
N+ +I SSSSSSS + AT S H++ +C V L N SSS
Sbjct: 68 NRPLIPKPSSSSSSSSICRATLSVRRISTTQPHLSTSCDVANFDPLGINSSSS 226
>TC77494 similar to SP|P21421|RPOB_PLAFA DNA-directed RNA polymerase beta
chain (EC 2.7.7.6). {Plasmodium falciparum}, partial
(1%)
Length = 943
Score = 27.7 bits (60), Expect = 1.7
Identities = 29/122 (23%), Positives = 50/122 (40%), Gaps = 13/122 (10%)
Frame = +2
Query: 3 TSLFRRLAPLLAARFQRNQNIISSFS-----SSSSSSVLHHATSSPSSPDVEPVHITENC 57
TSLF L A F N+ ++ S SS ++L SS + + ++ E
Sbjct: 359 TSLFTNLIDNNAQNFINNEESFTNESYLPSTSSEMDTMLSKLISSNNGWNNSEENLQEFD 538
Query: 58 VRKMKE--------LDANESSSGGKMLRLSVETGGCSGFQYAFNLDDRFNSDDRVFEKEG 109
V+ +K+ + N S G +++ G +GF +F + SDD K G
Sbjct: 539 VKPVKQEIGESVAQQNGNYSYGGSELIYQGFSNGSGNGFYGSFGGVNTMESDDSNQSKIG 718
Query: 110 IK 111
++
Sbjct: 719 VR 724
>AW695540 similar to GP|22597166|gb| RING-H2 finger protein {Glycine max},
partial (33%)
Length = 619
Score = 27.3 bits (59), Expect = 2.2
Identities = 24/72 (33%), Positives = 30/72 (41%)
Frame = +1
Query: 16 RFQRNQNIISSFSSSSSSSVLHHATSSPSSPDVEPVHITENCVRKMKELDANESSSGGKM 75
R RN +I SSF SS SS HH S D P VR+++ + G+
Sbjct: 40 RLGRN-DIDSSFHSSPSSDFHHHHRLHDSDDDDSPRIRRHYNVRRLRHHERVSERFDGRY 216
Query: 76 LRLSVETGGCSG 87
R V G SG
Sbjct: 217 RRSLVNEGVESG 252
>TC93205 similar to PIR|A86309|A86309 hypothetical protein AAD50008.1
[imported] - Arabidopsis thaliana, partial (7%)
Length = 689
Score = 27.3 bits (59), Expect = 2.2
Identities = 14/22 (63%), Positives = 15/22 (67%)
Frame = +3
Query: 25 SSFSSSSSSSVLHHATSSPSSP 46
SSFSSSSS S + TS P SP
Sbjct: 381 SSFSSSSSQSPISVVTSPPHSP 446
>TC78787 similar to GP|13569996|gb|AAK31280.1 putative Myb-related protein
{Oryza sativa}, partial (47%)
Length = 1438
Score = 27.3 bits (59), Expect = 2.2
Identities = 14/33 (42%), Positives = 20/33 (60%)
Frame = +2
Query: 20 NQNIISSFSSSSSSSVLHHATSSPSSPDVEPVH 52
N N+ + SSSSSS+ ++ +P SP EP H
Sbjct: 260 NLNLAALHHSSSSSSLNPGSSLNPGSPCFEPPH 358
>TC77056 similar to GP|15982817|gb|AAL09756.1 At2g20760/F5H14.27
{Arabidopsis thaliana}, partial (43%)
Length = 1228
Score = 27.3 bits (59), Expect = 2.2
Identities = 18/46 (39%), Positives = 22/46 (47%), Gaps = 16/46 (34%)
Frame = +1
Query: 27 FSSSSSSSV-LHHATSSPSSPDV---------------EPVHITEN 56
FSS S+ V + H T++P SPDV EPVH EN
Sbjct: 139 FSSYSAEDVPVDHTTAAPESPDVFGFSDQDPSYAQSPFEPVHAMEN 276
>TC88396 weakly similar to GP|18616497|emb|CAD22847. unnamed protein product
{Triticum aestivum}, partial (32%)
Length = 1399
Score = 27.3 bits (59), Expect = 2.2
Identities = 12/29 (41%), Positives = 22/29 (75%), Gaps = 1/29 (3%)
Frame = +2
Query: 41 SSPSSPDVEPVHITENCVRK-MKELDANE 68
S+P S +VEPV +++ +RK +K LD+++
Sbjct: 95 STPDSMEVEPVIVSDKSIRKILKALDSDD 181
>TC91850 similar to GP|22597166|gb|AAN03470.1 RING-H2 finger protein
{Glycine max}, partial (48%)
Length = 1054
Score = 27.3 bits (59), Expect = 2.2
Identities = 24/72 (33%), Positives = 30/72 (41%)
Frame = +2
Query: 16 RFQRNQNIISSFSSSSSSSVLHHATSSPSSPDVEPVHITENCVRKMKELDANESSSGGKM 75
R RN +I SSF SS SS HH S D P VR+++ + G+
Sbjct: 50 RLGRN-DIDSSFHSSPSSDFHHHHRLHDSDDDDSPRIRRHYNVRRLRHHERVSERFDGRY 226
Query: 76 LRLSVETGGCSG 87
R V G SG
Sbjct: 227 RRSLVNEGVESG 262
>TC82753 similar to GP|9758920|dbj|BAB09457.1
gb|AAD25781.1~gene_id:MXI22.10~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (14%)
Length = 816
Score = 26.9 bits (58), Expect = 2.9
Identities = 22/62 (35%), Positives = 27/62 (43%)
Frame = -2
Query: 17 FQRNQNIISSFSSSSSSSVLHHATSSPSSPDVEPVHITENCVRKMKELDANESSSGGKML 76
F N S SSSSSSS L SSP V P V K+K + + S +L
Sbjct: 191 FSAASNRSRSSSSSSSSSDLEDLESSPLIRSVIPESRLSK-VEKIKSMSSRSSLGAWGLL 15
Query: 77 RL 78
R+
Sbjct: 14 RI 9
>TC82350 similar to GP|9837385|gb|AAG00554.1| retinitis pigmentosa GTPase
regulator-like protein {Takifugu rubripes}, partial (2%)
Length = 759
Score = 26.9 bits (58), Expect = 2.9
Identities = 18/36 (50%), Positives = 20/36 (55%)
Frame = -2
Query: 26 SFSSSSSSSVLHHATSSPSSPDVEPVHITENCVRKM 61
SFSSSSSSS A+SS S PV T CV +
Sbjct: 203 SFSSSSSSSSFSDASSSSS-----PVSSTPYCVSSL 111
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.131 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,098,031
Number of Sequences: 36976
Number of extensions: 62272
Number of successful extensions: 906
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 671
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 795
length of query: 145
length of database: 9,014,727
effective HSP length: 87
effective length of query: 58
effective length of database: 5,797,815
effective search space: 336273270
effective search space used: 336273270
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 53 (25.0 bits)
Medicago: description of AC147007.11


