

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147002.5 + phase: 0
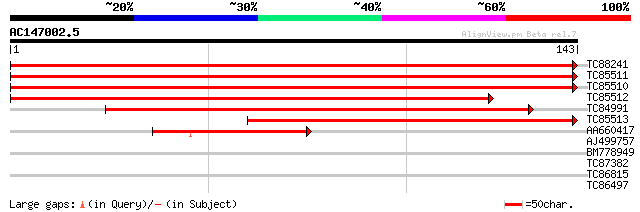
(143 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88241 similar to GP|11036950|gb|AAG27431.1 QM-like protein {El... 294 7e-81
TC85511 homologue to SP|Q9M5M7|RL10_EUPES 60S ribosomal protein ... 267 9e-73
TC85510 homologue to SP|Q9M5M7|RL10_EUPES 60S ribosomal protein ... 266 2e-72
TC85512 homologue to SP|P45621|GSA_SOYBN Glutamate-1-semialdehyd... 226 2e-60
TC84991 similar to PIR|A57296|A57296 ribosomal protein L10.e cy... 149 3e-37
TC85513 weakly similar to PIR|A96615|A96615 probable dirigent pr... 144 9e-36
AA660417 homologue to SP|P93847|RL10_ 60S ribosomal protein L10 ... 51 1e-07
AJ499757 similar to PIR|T02711|T02 probable calmodulin [imported... 28 1.6
BM778949 27 2.1
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 27 2.1
TC86815 similar to GP|15289792|dbj|BAB63491. contains ESTs AU100... 26 6.2
TC86497 similar to GP|21535793|emb|CAC85725. putative carbamoyl ... 25 8.1
>TC88241 similar to GP|11036950|gb|AAG27431.1 QM-like protein {Elaeis
guineensis}, partial (95%)
Length = 954
Score = 294 bits (753), Expect = 7e-81
Identities = 143/143 (100%), Positives = 143/143 (100%)
Frame = +1
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL
Sbjct: 307 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 486
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV
Sbjct: 487 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 666
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAKVLGCHGPLANRQPGRAFLPA
Sbjct: 667 NAKVLGCHGPLANRQPGRAFLPA 735
>TC85511 homologue to SP|Q9M5M7|RL10_EUPES 60S ribosomal protein L10. [Leafy
spurge] {Euphorbia esula}, partial (97%)
Length = 891
Score = 267 bits (683), Expect = 9e-73
Identities = 129/143 (90%), Positives = 134/143 (93%)
Frame = +2
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
MSKFAGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP G CARV+IGQVL
Sbjct: 281 MSKFAGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGVCARVAIGQVL 460
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK N HAQEALRRAKFKFPGRQKIIVSRKWGFTK++ AEYLKLKSENRI+PDGV
Sbjct: 461 LSVRCKDNNSHHAQEALRRAKFKFPGRQKIIVSRKWGFTKINRAEYLKLKSENRIMPDGV 640
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAK+LGCHGPLANRQPGRAFL A
Sbjct: 641 NAKLLGCHGPLANRQPGRAFLNA 709
>TC85510 homologue to SP|Q9M5M7|RL10_EUPES 60S ribosomal protein L10. [Leafy
spurge] {Euphorbia esula}, partial (97%)
Length = 904
Score = 266 bits (681), Expect = 2e-72
Identities = 128/143 (89%), Positives = 134/143 (93%)
Frame = +2
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
MSKFAGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP G CARV+IGQVL
Sbjct: 269 MSKFAGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGVCARVAIGQVL 448
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK N HAQEALRRAKFKFPGRQKIIVSRKWGFTK++ A+YLKLKSENRI+PDGV
Sbjct: 449 LSVRCKDNNSHHAQEALRRAKFKFPGRQKIIVSRKWGFTKINRADYLKLKSENRIMPDGV 628
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAK+LGCHGPLANRQPGRAFL A
Sbjct: 629 NAKLLGCHGPLANRQPGRAFLTA 697
>TC85512 homologue to SP|P45621|GSA_SOYBN Glutamate-1-semialdehyde 2
1-aminomutase chloroplast precursor (EC 5.4.3.8) (GSA),
partial (91%)
Length = 2272
Score = 226 bits (576), Expect = 2e-60
Identities = 109/122 (89%), Positives = 114/122 (93%)
Frame = -2
Query: 1 MSKFAGKDTFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVL 60
MSKFAGKD FHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKP G CARV+IGQVL
Sbjct: 375 MSKFAGKDAFHLRVRVHPFHVLRINKMLSCAGADRLQTGMRGAFGKPQGVCARVAIGQVL 196
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVRCK N HAQEALRRAKFKFPGRQKIIVSRKWGFTK++ A+YLKLKSENRI+PDGV
Sbjct: 195 LSVRCKDNNSHHAQEALRRAKFKFPGRQKIIVSRKWGFTKINRADYLKLKSENRIMPDGV 16
Query: 121 NA 122
NA
Sbjct: 15 NA 10
>TC84991 similar to PIR|A57296|A57296 ribosomal protein L10.e cytosolic -
yeast (Saccharomyces cerevisiae), partial (48%)
Length = 432
Score = 149 bits (377), Expect = 3e-37
Identities = 73/108 (67%), Positives = 82/108 (75%)
Frame = +2
Query: 25 NKMLSCAGADRLQTGMRGAFGKPLGTCARVSIGQVLLSVRCKSGNGDHAQEALRRAKFKF 84
NKMLSCAGADRLQTGMRGAFGKP GT ARV IGQ++ SVR K N EALRRAK+KF
Sbjct: 2 NKMLSCAGADRLQTGMRGAFGKPTGTVARVDIGQIIFSVRTKDNNKPTVIEALRRAKYKF 181
Query: 85 PGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGVNAKVLGCHGPLA 132
PGRQKIIVS+KWGFTKLS EY++ + + PDG K L HGPL+
Sbjct: 182 PGRQKIIVSKKWGFTKLSREEYMEKRQNGLLQPDGAYVKYLSHHGPLS 325
>TC85513 weakly similar to PIR|A96615|A96615 probable dirigent protein
T18I24.8 [imported] - Arabidopsis thaliana, partial
(58%)
Length = 1098
Score = 144 bits (364), Expect = 9e-36
Identities = 70/83 (84%), Positives = 75/83 (90%)
Frame = +1
Query: 61 LSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
LSVR K N HAQ+ALRRAKFKFPGRQK IVSRKWGFTK++ AEYLKLKSENRI+PDGV
Sbjct: 1 LSVRGKDNNRRHAQDALRRAKFKFPGRQKSIVSRKWGFTKINRAEYLKLKSENRIMPDGV 180
Query: 121 NAKVLGCHGPLANRQPGRAFLPA 143
NAK+LGCHGPLANRQPGRAFL A
Sbjct: 181 NAKLLGCHGPLANRQPGRAFLNA 249
>AA660417 homologue to SP|P93847|RL10_ 60S ribosomal protein L10 (EQM).
[Eggplant Aubergine] {Solanum melongena}, partial (37%)
Length = 686
Score = 51.2 bits (121), Expect = 1e-07
Identities = 25/41 (60%), Positives = 29/41 (69%), Gaps = 1/41 (2%)
Frame = +1
Query: 37 QTGMRGAF-GKPLGTCARVSIGQVLLSVRCKSGNGDHAQEA 76
Q GM+G P G CA+++IGQVLLSVRCK N HAQEA
Sbjct: 349 QLGMKGLLLESPQGVCAKMAIGQVLLSVRCKDNNSHHAQEA 471
>AJ499757 similar to PIR|T02711|T02 probable calmodulin [imported] -
Arabidopsis thaliana, partial (3%)
Length = 409
Score = 27.7 bits (60), Expect = 1.6
Identities = 17/50 (34%), Positives = 24/50 (48%)
Frame = +1
Query: 71 DHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGV 120
DH AK + P +I S+ WG + +SH L +K I+P GV
Sbjct: 175 DHLSFHQSSAKQQMPN---LITSKSWGASNVSHPHQLDIKYP--IIPPGV 309
>BM778949
Length = 166
Score = 27.3 bits (59), Expect = 2.1
Identities = 14/35 (40%), Positives = 19/35 (54%)
Frame = +3
Query: 44 FGKPLGTCARVSIGQVLLSVRCKSGNGDHAQEALR 78
FG C +VS+ SVRC +GNG A E ++
Sbjct: 3 FGSNRTHCLKVSV----FSVRCSTGNGRKAMEKIQ 95
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 27.3 bits (59), Expect = 2.1
Identities = 15/51 (29%), Positives = 27/51 (52%), Gaps = 1/51 (1%)
Frame = -3
Query: 60 LLSVRCKSGNGDHAQEALRRAKFKFPGRQKIIVSR-KWGFTKLSHAEYLKL 109
L+ + SGN ++ L + FP + ++VSR KW +++S + L L
Sbjct: 382 LVKIVVVSGNFVGGEDLLTKGFLHFPNKMTVLVSRTKWPQSEVSQRQLLAL 230
>TC86815 similar to GP|15289792|dbj|BAB63491. contains ESTs AU100716(C12248)
AU029967(E50373) C26395(C12248)~hypothetical
protein~similar to, partial (47%)
Length = 997
Score = 25.8 bits (55), Expect = 6.2
Identities = 15/61 (24%), Positives = 27/61 (43%)
Frame = -3
Query: 64 RCKSGNGDHAQEALRRAKFKFPGRQKIIVSRKWGFTKLSHAEYLKLKSENRIVPDGVNAK 123
+C+ GNG H Q++ + K+ S+ WG + H L ++ I D +N
Sbjct: 242 KCREGNGHHQQQSGSFVQI------KVCKSKNWGPVEEGHG---LLPCQHLISHDAINTT 90
Query: 124 V 124
+
Sbjct: 89 I 87
>TC86497 similar to GP|21535793|emb|CAC85725. putative carbamoyl phosphate
synthase small subunit {Nicotiana tabacum}, partial
(64%)
Length = 1199
Score = 25.4 bits (54), Expect = 8.1
Identities = 13/25 (52%), Positives = 16/25 (64%)
Frame = +2
Query: 47 PLGTCARVSIGQVLLSVRCKSGNGD 71
P T A+VS+ SVRC SGNG+
Sbjct: 248 PPHTSAKVSV----FSVRCSSGNGE 310
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.140 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,392,561
Number of Sequences: 36976
Number of extensions: 53934
Number of successful extensions: 299
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 299
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 299
length of query: 143
length of database: 9,014,727
effective HSP length: 87
effective length of query: 56
effective length of database: 5,797,815
effective search space: 324677640
effective search space used: 324677640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 53 (25.0 bits)
Medicago: description of AC147002.5


