

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147000.7 - phase: 0
(1185 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
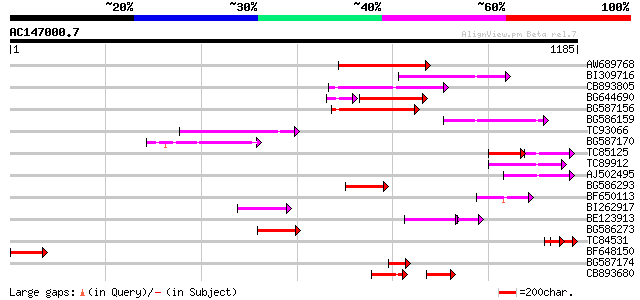
Score E
Sequences producing significant alignments: (bits) Value
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 184 2e-46
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 163 4e-40
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 154 1e-37
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 125 3e-35
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 143 4e-34
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 137 3e-32
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 122 6e-28
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 117 2e-26
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 71 2e-23
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 96 8e-20
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 95 1e-19
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 89 1e-17
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 87 4e-17
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 84 2e-16
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 66 5e-16
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 82 2e-15
TC84531 77 4e-14
BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x pa... 74 3e-13
BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse ... 74 4e-13
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 74 4e-13
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 184 bits (466), Expect = 2e-46
Identities = 86/192 (44%), Positives = 127/192 (65%)
Frame = +1
Query: 687 WQEAINDEMDSLMSNETWHLTDLPPGCKTIGCKWILKKKLKPDGSIDKYKARLVAKGFRQ 746
W +A+ E +L+ N+TW L LPP K IGCKW+ + K PDGS++K+KARLVAKGF Q
Sbjct: 97 WLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVAKGFSQ 276
Query: 747 RENVDFFDTYSPVTRITSIRVLISLAAIHNLIVHQMDVKTAFLNGELEEEIYMDQPEGFV 806
D+ +T+SPV + +IR+++++A + + Q+D+ AFLNG L+EE+YM QP+GF
Sbjct: 277 TLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQPQGFE 456
Query: 807 IHGQENKVCKLDKSLYGLKQAPKQWHEKFDNLMIENEFKVNESDKCIYSKYENNTCTIIC 866
++ VCKL+KSLYGLKQAP+ W+E + I+ F + D + +N C +
Sbjct: 457 A-ANKSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYNQNGACIYLX 633
Query: 867 LYVDDLLIFGSN 878
+YVDD+LI GS+
Sbjct: 634 IYVDDILITGSS 669
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 163 bits (412), Expect = 4e-40
Identities = 88/237 (37%), Positives = 137/237 (57%), Gaps = 2/237 (0%)
Frame = +2
Query: 813 KVCKLDKSLYGLKQAPKQWHEKFDNLMIENEFKVNESDKCIYSKYENNTCTIICLYVDDL 872
KVC+L KS+YGLKQA +QW+ K +I + + SD +++K+++++ T + +YVDD+
Sbjct: 17 KVCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDI 196
Query: 873 LIFGSNLNAIKDVKSLLCHNFDMKDLGKADVILGIKITRTDNGISLNQSHYVEKILRKYN 932
++ G++++ I+ VK L F +KDLG LG+++ R+ GI LNQ Y ++L
Sbjct: 197 VLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSG 376
Query: 933 YFYCKPASTPCDPSVKLFKNTGDSVR-QTEYASIIGSLRYATDCTRPDISYAVGLLCKFT 991
K TP D S+KL + +T+Y +IG L Y T TRPDIS+AV L +F
Sbjct: 377 NLAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLT-TTRPDISFAVQQLSQFV 553
Query: 992 SRPSMEHWQAIERVMRYLKKTMTLGLHYQRYPAV-LEGYSDADWNNLSDDSKATSGY 1047
S+P H+QA RV++YLK GL Y + L ++D+DW K+ +GY
Sbjct: 554 SKPQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGY 724
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 154 bits (390), Expect = 1e-37
Identities = 87/254 (34%), Positives = 147/254 (57%), Gaps = 4/254 (1%)
Frame = +3
Query: 667 TIEEDPSSIKEALSSIDADLWQEAINDEMDSLMSNETWHLTDLPPGCKTIGCKWILKKKL 726
T+ DP++ +EA+ S + W+ ++N+EM++ N TW LTDL G KTIG KWI K KL
Sbjct: 27 TMTSDPTTFEEAVKS---EKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKL 197
Query: 727 KPDGSIDKYKARLVAKGFRQRENVDFFDTYSPVTRITSIRVLISLAA---IHNLIVHQMD 783
+G I+KYKARLVAKG+ Q+ VD+ + ++PV R +IR++I+LAA + + M
Sbjct: 198 NENGEIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAAQIKRDGVCIS*M* 377
Query: 784 VKTAFLNGELEEEIYMDQPEGFVIHGQENKVCKLDKSLYGLKQAPKQWHEKFDNLMIENE 843
+ + + + + ++ + + ++ ++LYGLKQAP+ W+ + + +
Sbjct: 378 KAHSCMEN*MRKFLLINHR----VM*RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTKEG 545
Query: 844 FKVNESDKCIYSKY-ENNTCTIICLYVDDLLIFGSNLNAIKDVKSLLCHNFDMKDLGKAD 902
F+ + ++ K E II LYVDDL+ G++ N ++ K + F+M DLGK
Sbjct: 546 FEKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKMH 725
Query: 903 VILGIKITRTDNGI 916
LG+++T+ + GI
Sbjct: 726 YFLGVEVTQNEKGI 767
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 125 bits (314), Expect(2) = 3e-35
Identities = 58/141 (41%), Positives = 95/141 (67%)
Frame = -2
Query: 732 IDKYKARLVAKGFRQRENVDFFDTYSPVTRITSIRVLISLAAIHNLIVHQMDVKTAFLNG 791
I + K++LV +G+ Q+E +D+ + +SPV R+ +IR+LI+ AA ++QMDVK+AF+NG
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 792 ELEEEIYMDQPEGFVIHGQENKVCKLDKSLYGLKQAPKQWHEKFDNLMIENEFKVNESDK 851
+L+EE+++ QP GF N V +L+K+LYGLKQAP+ W+E+ +++N FK + D
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 852 CIYSKYENNTCTIICLYVDDL 872
++ II +YVDD+
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 42.7 bits (99), Expect(2) = 3e-35
Identities = 18/65 (27%), Positives = 34/65 (51%)
Frame = -3
Query: 663 YVAYTIEEDPSSIKEALSSIDADLWQEAINDEMDSLMSNETWHLTDLPPGCKTIGCKWIL 722
+ A+ +P ++KEAL D W ++ +E+ ++ W+L P G IG +W+
Sbjct: 612 FSAFISSIEPKNVKEALRDAD---WINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVF 442
Query: 723 KKKLK 727
+ KL+
Sbjct: 441 RNKLE 427
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 143 bits (360), Expect = 4e-34
Identities = 69/184 (37%), Positives = 113/184 (60%)
Frame = -1
Query: 672 PSSIKEALSSIDADLWQEAINDEMDSLMSNETWHLTDLPPGCKTIGCKWILKKKLKPDGS 731
P S +EA+ + W+E++ E +++ N+TW+ ++LP G K + +WI K K DGS
Sbjct: 558 PRSYEEAMEDKE---WKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWIFTIKYKADGS 388
Query: 732 IDKYKARLVAKGFRQRENVDFFDTYSPVTRITSIRVLISLAAIHNLIVHQMDVKTAFLNG 791
I++ K RLVA+GF D+ +T++PV ++ +IR+++SLA + QMDVK AFL G
Sbjct: 387 IERKKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQMDVKNAFLQG 208
Query: 792 ELEEEIYMDQPEGFVIHGQENKVCKLDKSLYGLKQAPKQWHEKFDNLMIENEFKVNESDK 851
ELE+E+YM P G + V +L K++YGLKQ+P+ W+ K + F+ +E D
Sbjct: 207 ELEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNGRGFRKSELDH 28
Query: 852 CIYS 855
+++
Sbjct: 27 TLFT 16
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 137 bits (344), Expect = 3e-32
Identities = 75/221 (33%), Positives = 128/221 (56%), Gaps = 2/221 (0%)
Frame = +1
Query: 907 IKITRTDNGISLNQSHYVEKILRKYNYFYCKPASTPCDPSVKLFKN-TGDSVRQTEYASI 965
+++ + + GI + Q YV +L ++ + P P KL K+ G V T+Y I
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 966 IGSLRYATDCTRPDISYAVGLLCKFTSRPSMEHWQAIERVMRYLKKTMTLGLHYQRYPAV 1025
+G L Y TRPD+ Y + L+ +F + P+ H A++RV+RYL T+ LG+ Y+R +
Sbjct: 181 VGCLMYLA-ATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSE 357
Query: 1026 -LEGYSDADWNNLSDDSKATSGYIFSIAGGAVSWKSKKQTILAQSTMESEMIALAAASEE 1084
LE Y+D+D+ DD K+TSGY+F ++ GAVSW SKKQ ++ ST ++E IA A + +
Sbjct: 358 KLEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQ 537
Query: 1085 ASWLRCLLSEIPLWERPLPAVLIHCDSTAAIAKIENRYYNG 1125
+ W+R +L ++ + ++ ++CD+ + I +N +G
Sbjct: 538 SVWMRRVLEKLGYTQS--GSITMYCDNNSTIKLSKNPVLHG 654
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 122 bits (307), Expect = 6e-28
Identities = 90/261 (34%), Positives = 123/261 (46%), Gaps = 10/261 (3%)
Frame = +1
Query: 356 EKCQFCSQAKI----NKESHKSVTRITEPF-ELIHSDLCELDGNLTRNGKRYFITFIDDC 410
+K +FC K S + T T+ + IHSDL + G+RY +T IDD
Sbjct: 7 DKLEFCKHLLFFGNRKKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDF 186
Query: 411 SDYTHVYLMRNKNEALDIFKQYVKEIENQFNIRIKRFRSDRGTEYGSHIFNEYYKELGII 470
VY +R KNE FK++ +E Q +K+ +D E+ S FNE+ GI
Sbjct: 187 PRKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIA 366
Query: 471 HETTAPYSPEMNGKAERKNRTFTELVVATMLNSGA--APHWWGEILLTVCYVLNRVPKTK 528
T P +P+ NG AER RT E + N+G W E T C+++NR P +
Sbjct: 367 RHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSA 546
Query: 529 NKIS-PYEILKKRQPNLSYFRTWGCLAYVRKPDPKRVKLASRAYECAFIGYALNSKAYRF 587
P +I + S R +GC AY D KLA RA EC F+ YA SK YR
Sbjct: 547 LDFKVPEDIWSGNLVDYSNLRIFGCPAYALVNDG---KLAPRAGECIFLSYASESKGYRL 717
Query: 588 Y--DLKSKTIIESNDVDFYEN 606
+ D KS+ +I S DV F E+
Sbjct: 718 WCSDPKSQKLILSRDVTFNED 780
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 117 bits (294), Expect = 2e-26
Identities = 71/249 (28%), Positives = 127/249 (50%), Gaps = 10/249 (4%)
Frame = -3
Query: 287 ITKNGIFVGKGYATDGMFKLNIDMNKISSSAYMLCDFN---------IWHSRLCHVNKRI 337
+ ++ +G+G ++ L K+ + C F +WH+RL H + R
Sbjct: 713 VIESSQLIGEGVTKGDLYMLE----KLDPVSNYKCSFTSSSSLNKDALWHARLGHPHGRA 546
Query: 338 ISNMSGLGLIPKISLNDFEKCQFCSQAKINKESHKSVTRITEP-FELIHSDLCELDGNLT 396
++ M +P + + + C+ C K K + + E F+LI++DL +L+
Sbjct: 545 LNLM-----LPGVVFEN-KNCEACILGKHCKNVFPRTSTVYENCFDLIYTDLWTAP-SLS 387
Query: 397 RNGKRYFITFIDDCSDYTHVYLMRNKNEALDIFKQYVKEIENQFNIRIKRFRSDRGTEYG 456
R+ +YF+TFID+ S YT + L+ +K+ +D FK + + N ++ +IK RSD G EY
Sbjct: 386 RDNHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKNFQAYVTNHYHAKIKILRSDNGGEYT 207
Query: 457 SHIFNEYYKELGIIHETTAPYSPEMNGKAERKNRTFTELVVATMLNSGAAPHWWGEILLT 516
S+ F + GI+H+T+ PY+P+ NG A+RKN+ E+ + M + + T
Sbjct: 206 SYAFKSHLDHHGILHQTSCPYTPQQNGVAKRKNKHLMEVARSLMFQANVS---------T 54
Query: 517 VCYVLNRVP 525
CY++N +P
Sbjct: 53 ACYLINWIP 27
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 71.2 bits (173), Expect(2) = 2e-23
Identities = 33/76 (43%), Positives = 52/76 (68%)
Frame = +1
Query: 1002 IERVMRYLKKTMTLGLHYQRYPAVLEGYSDADWNNLSDDSKATSGYIFSIAGGAVSWKSK 1061
++R+MRY+K T + + + + GY D+D+ D K+T+GY+F++AGGAVSW SK
Sbjct: 1 VKRIMRYIKGTSGVAVCFGGSELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLSK 180
Query: 1062 KQTILAQSTMESEMIA 1077
QT++A ST E+E +A
Sbjct: 181 LQTVVALSTTEAEYMA 228
Score = 57.8 bits (138), Expect(2) = 2e-23
Identities = 30/104 (28%), Positives = 58/104 (54%)
Frame = +3
Query: 1077 ALAAASEEASWLRCLLSEIPLWERPLPAVLIHCDSTAAIAKIENRYYNGKRRQIRRKHST 1136
+L A +EA W++ L+ E+ + + ++CDS +A+ N ++ + + I ++
Sbjct: 228 SLPQACKEAIWMQRLMEELGHKQEQ---ITVYCDSQSALHIARNPAFHSRTKHIGIQYHF 398
Query: 1137 IREYLSNGTVRVDFVRTNENLADPLTKGLNREKVANTSSRMGLM 1180
+RE + G+V + + TN+NLAD +TK +N +K S GL+
Sbjct: 399 VREVVEEGSVDMQKIHTNDNLADAMTKSINTDKFIWCRSSYGLL 530
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 95.9 bits (237), Expect = 8e-20
Identities = 52/167 (31%), Positives = 96/167 (57%), Gaps = 3/167 (1%)
Frame = +1
Query: 1000 QAIERVMRYLKKTMTLGLHYQRYPA---VLEGYSDADWNNLSDDSKATSGYIFSIAGGAV 1056
QA++ V++YL +++ L Y + LEGY DAD+ D K+ SG++F++ G +
Sbjct: 1 QALKWVLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTI 180
Query: 1057 SWKSKKQTILAQSTMESEMIALAAASEEASWLRCLLSEIPLWERPLPAVLIHCDSTAAIA 1116
SWK+ +Q+++ ST ++E IA ++A WL+ ++ E+ + + V IHCDS +AI
Sbjct: 181 SWKANQQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGITQE---YVKIHCDSQSAIH 351
Query: 1117 KIENRYYNGKRRQIRRKHSTIREYLSNGTVRVDFVRTNENLADPLTK 1163
++ Y+ + + I + IR+ + + + V+ + + EN AD TK
Sbjct: 352 LANHQVYHERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTK 492
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 95.1 bits (235), Expect = 1e-19
Identities = 53/149 (35%), Positives = 85/149 (56%)
Frame = +2
Query: 1032 ADWNNLSDDSKATSGYIFSIAGGAVSWKSKKQTILAQSTMESEMIALAAASEEASWLRCL 1091
+DW ++ K+TSGY F + GA+SW SKKQ ++A ST E+E IA + + + WLR +
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 1092 LSEIPLWERPLPAVLIHCDSTAAIAKIENRYYNGKRRQIRRKHSTIREYLSNGTVRVDFV 1151
L E+ E+ P I+CD+ +AIA +N ++G+ + I + IRE ++ V +++
Sbjct: 182 L-EVMHHEQNTP-TKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYC 355
Query: 1152 RTNENLADPLTKGLNREKVANTSSRMGLM 1180
T E +AD TK L E +G+M
Sbjct: 356 PTEEKIADIFTKPLKIESFYKLKKMLGMM 442
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 89.0 bits (219), Expect = 1e-17
Identities = 43/90 (47%), Positives = 62/90 (68%)
Frame = +2
Query: 702 ETWHLTDLPPGCKTIGCKWILKKKLKPDGSIDKYKARLVAKGFRQRENVDFFDTYSPVTR 761
+T L P G K IG +WI K K DG++ KYKARLVAKG+ +++ +DF + ++PV R
Sbjct: 53 QTLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPVVR 232
Query: 762 ITSIRVLISLAAIHNLIVHQMDVKTAFLNG 791
I +I +L++LAA + +H +DVK AFLNG
Sbjct: 233 IETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 87.0 bits (214), Expect = 4e-17
Identities = 51/126 (40%), Positives = 72/126 (56%), Gaps = 5/126 (3%)
Frame = +1
Query: 975 CTRPDISYAVGLLCKFTSRPSMEHWQAIERVMRYLKKTMTLGLHYQRYPAVLEGY----- 1029
C RPDI Y+V ++ KF P H A R++RY++ TM GL + Y A E Y
Sbjct: 115 C*RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFP-YGAKSEVYELICY 291
Query: 1030 SDADWNNLSDDSKATSGYIFSIAGGAVSWKSKKQTILAQSTMESEMIALAAASEEASWLR 1089
SD+DW D ++TSGY+F A+SW +KKQ I A S+ E+E IA A+ +A WL
Sbjct: 292 SDSDW---CGDRRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLD 462
Query: 1090 CLLSEI 1095
++ E+
Sbjct: 463 SVIKEL 480
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 84.3 bits (207), Expect = 2e-16
Identities = 43/114 (37%), Positives = 63/114 (54%), Gaps = 1/114 (0%)
Frame = +1
Query: 477 YSPEMNGKAERKNRTFTELVVATMLNSGAAPHWWGEILLTVCYVLNRVPKTKNKI-SPYE 535
++P+ NG AER NRT E A + +G A +W E + T CYV+NR P T + +P E
Sbjct: 82 HTPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPME 261
Query: 536 ILKKRQPNLSYFRTWGCLAYVRKPDPKRVKLASRAYECAFIGYALNSKAYRFYD 589
+ K + + S +GC YV +R KL ++ +C F+GYA N K Y +D
Sbjct: 262 MWKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGYXLWD 423
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 66.2 bits (160), Expect(2) = 5e-16
Identities = 34/118 (28%), Positives = 65/118 (54%), Gaps = 1/118 (0%)
Frame = +1
Query: 826 QAPKQWHEKFDNLMIENEFKVNESDKCIYSKYENNTC-TIICLYVDDLLIFGSNLNAIKD 884
Q+P+ W ++F ++ + + ++D ++ K+ + I+ +YVDD+ + G + IK
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 885 VKSLLCHNFDMKDLGKADVILGIKITRTDNGISLNQSHYVEKILRKYNYFYCKPASTP 942
+K+LL F++KDLG LG+++ R G S++Q YV +L++ CK P
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Score = 37.4 bits (85), Expect(2) = 5e-16
Identities = 22/55 (40%), Positives = 32/55 (58%), Gaps = 1/55 (1%)
Frame = +2
Query: 937 KPASTPCDPSVKL-FKNTGDSVRQTEYASIIGSLRYATDCTRPDISYAVGLLCKF 990
KP+ TP D +VKL + G V + Y ++G L Y + TRPDIS+ V + +F
Sbjct: 338 KPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSH-TRPDISFVVCTMSQF 499
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 81.6 bits (200), Expect = 2e-15
Identities = 36/91 (39%), Positives = 59/91 (64%), Gaps = 1/91 (1%)
Frame = -2
Query: 518 CYVLNRVP-KTKNKISPYEILKKRQPNLSYFRTWGCLAYVRKPDPKRVKLASRAYECAFI 576
CY++NR+P + +P+E+L +R+P+L+Y R +GCL YV P R KL +R+ + FI
Sbjct: 701 CYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMFI 522
Query: 577 GYALNSKAYRFYDLKSKTIIESNDVDFYENK 607
GY+ K Y+ YD +++ ++ S DV F E +
Sbjct: 521 GYSTTQKGYKCYDPEARRVLVSRDVKFIEER 429
>TC84531
Length = 655
Score = 77.0 bits (188), Expect = 4e-14
Identities = 37/41 (90%), Positives = 38/41 (92%)
Frame = -3
Query: 1118 IENRYYNGKRRQIRRKHSTIREYLSNGTVRVDFVRTNENLA 1158
IENRYYNGKRRQIRRKHSTIREYLSNGTVRVDFVRT L+
Sbjct: 653 IENRYYNGKRRQIRRKHSTIREYLSNGTVRVDFVRTK*KLS 531
Score = 54.3 bits (129), Expect = 3e-07
Identities = 32/57 (56%), Positives = 36/57 (63%), Gaps = 2/57 (3%)
Frame = -2
Query: 1131 RRKHSTIREYLSNGTVRVDFVRTNENLA--DPLTKGLNREKVANTSSRMGLMPIDHK 1185
+ +H + NG R RTN+ DPLTKGL REKVANTS RMGLMPIDHK
Sbjct: 609 KTQHDKRISFERNGKSRF---RTNKMKT*PDPLTKGLTREKVANTSIRMGLMPIDHK 448
>BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x paradisi},
partial (3%)
Length = 658
Score = 73.9 bits (180), Expect = 3e-13
Identities = 36/78 (46%), Positives = 51/78 (65%)
Frame = +2
Query: 1 MSDDKSVEAQSHELQQIAHEIIAEGMALPEQFQIAVIIDKLPPAWKDFKSLLRHKTKEFS 60
M D+KSV Q +E+++I + M + E ++ IIDKLPP+WKDFK ++HK ++ S
Sbjct: 359 MVDNKSVMEQLYEIERILNNYKQHNMNMDETIIVSSIIDKLPPSWKDFKRTMKHKKEDIS 538
Query: 61 LESLITRLRIEEEARKQE 78
LE L LR+ EE RKQE
Sbjct: 539 LEQLGNHLRLXEEYRKQE 592
>BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse
transcriptase homolog - rape retrotransposon copia-like
(fragment), partial (84%)
Length = 249
Score = 73.6 bits (179), Expect = 4e-13
Identities = 31/46 (67%), Positives = 41/46 (88%)
Frame = -1
Query: 792 ELEEEIYMDQPEGFVIHGQENKVCKLDKSLYGLKQAPKQWHEKFDN 837
ELEE+IYM QPEGF+ G+E+ VCKL KSLYGLKQ+P+QW+++FD+
Sbjct: 249 ELEEKIYMTQPEGFLFPGKEDHVCKLRKSLYGLKQSPRQWYKRFDS 112
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 73.6 bits (179), Expect = 4e-13
Identities = 38/75 (50%), Positives = 51/75 (67%)
Frame = -2
Query: 756 YSPVTRITSIRVLISLAAIHNLIVHQMDVKTAFLNGELEEEIYMDQPEGFVIHGQENKVC 815
+ P+ ++ +I L+S+ AI NL + +DVKTAFL G+L E+IYM QPEGF V
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS*E-VGKMVG 378
Query: 816 KLDKSLYGLKQAPKQ 830
KL KS+YGLKQ P+Q
Sbjct: 377 KLKKSMYGLKQGPRQ 333
Score = 45.4 bits (106), Expect = 1e-04
Identities = 24/63 (38%), Positives = 39/63 (61%), Gaps = 2/63 (3%)
Frame = -3
Query: 872 LLIFGSNLNAIKDVKSLLCHNFDMKDLGKADVILG--IKITRTDNGISLNQSHYVEKILR 929
LL+ GSN++ IK++K+ DMKDLG A I+G I I + + L+Q Y+ ++L+
Sbjct: 271 LLVVGSNIDEIKNLKTRFSKEIDMKDLGPAKKIIGMQIMIDKQKGVL*LSQVEYITRVLQ 92
Query: 930 KYN 932
+N
Sbjct: 91 IFN 83
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.134 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,874,248
Number of Sequences: 36976
Number of extensions: 513011
Number of successful extensions: 2748
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 2678
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2729
length of query: 1185
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1078
effective length of database: 5,058,295
effective search space: 5452842010
effective search space used: 5452842010
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC147000.7


