

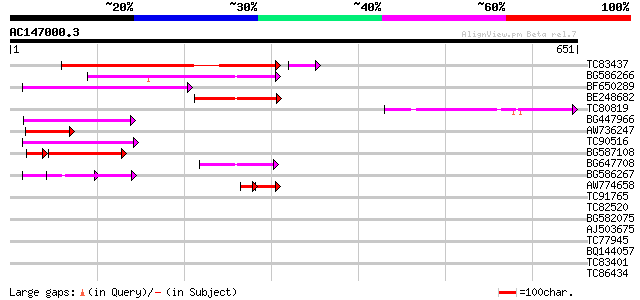
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147000.3 + phase: 0 /pseudo
(651 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imp... 204 5e-54
BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-li... 134 1e-31
BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgari... 132 3e-31
BE248682 similar to GP|18568269|gb putative gag-pol polyprotein ... 107 1e-23
TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non... 105 4e-23
BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [impo... 100 3e-21
AW736247 weakly similar to PIR|T14619|T146 reverse transcriptase... 79 4e-15
TC90516 78 1e-14
BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - ... 74 3e-14
BG647708 weakly similar to GP|13786450|gb| putative reverse tran... 72 9e-13
BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At... 62 9e-10
AW774658 similar to GP|2808681|emb| Hcr9-4B {Lycopersicon hirsut... 36 7e-06
TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative ret... 40 0.003
TC82520 32 0.47
BG582075 similar to PIR|T05242|T052 hypothetical protein F18A5.1... 32 0.77
AJ503675 similar to GP|21928168|gb| AT5g23040/MYJ24_3 {Arabidops... 31 1.7
TC77945 similar to SP|O24296|GSHY_PEA Phospholipid hydroperoxide... 30 2.2
BQ144057 similar to GP|1483567|emb| viral proteinase {Pseudorabi... 30 2.2
TC83401 homologue to GP|10441918|gb|AAG17236.1 unknown {Homo sap... 30 3.8
TC86434 similar to GP|2262100|gb|AAB63608.1| unknown protein {Ar... 29 5.0
>TC83437 weakly similar to PIR|D86384|D86384 unknown protein [imported] -
Arabidopsis thaliana, partial (6%)
Length = 951
Score = 204 bits (520), Expect(2) = 5e-54
Identities = 116/252 (46%), Positives = 158/252 (62%), Gaps = 1/252 (0%)
Frame = +2
Query: 60 QFLVDFHRNGKLIKGVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNV 119
+F+ +FHRN KL KG+NSTFIALIPKV+NPQRLND+ PISLVG LYK+L K+LANRLR V
Sbjct: 5 RFVSEFHRNRKLFKGINSTFIALIPKVDNPQRLNDFRPISLVGSLYKILGKLLANRLRVV 184
Query: 120 IGSVVSDSQSAFIKGN*ILDGILIANEVVDDARRMDKELLLFRVDFEKAYDSVDLKYLDM 179
IGSV+SD+QSAF+K IL+ + + + R +++ K ++ + + +
Sbjct: 185 IGSVISDAQSAFVKNRQILEMVFL*QMRLWMRLRN*RKIFCCLRWILKRLITLSIGLIWI 364
Query: 180 VM-VTMNFPTLWQKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEG 238
+ V M+F LW+KWI EC+ TA +VLVNG P
Sbjct: 365 LF*VGMSFLVLWRKWIKECVSTATTSVLVNGSPT-------------------------- 466
Query: 239 FNVLMKQMAGAQLFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEV 298
NVLMK + QLF + G V V ++HLQFA+DTL++ K+W N+R++RA L++F +
Sbjct: 467 -NVLMKSLVQTQLFTRYSFGVVNPVVVSHLQFANDTLLLETKNWANIRALRAALVIF*AM 643
Query: 299 SGLKVNFHKSML 310
SGLKVNFHKS L
Sbjct: 644 SGLKVNFHKSGL 679
Score = 25.4 bits (54), Expect(2) = 5e-54
Identities = 11/37 (29%), Positives = 17/37 (45%)
Frame = +1
Query: 321 GICIELSKREASFCLFGLTYWRGFSKIEFLETCCRPY 357
G C +L + + + Y FS +EFL T C +
Sbjct: 718 GFCFKLESGQGTIFVLRYAYRGKFSALEFLGTYCESH 828
>BG586266 similar to GP|7267666|em RNA-directed DNA polymerase-like protein
{Arabidopsis thaliana}, partial (18%)
Length = 789
Score = 134 bits (336), Expect = 1e-31
Identities = 74/225 (32%), Positives = 123/225 (53%), Gaps = 3/225 (1%)
Frame = -3
Query: 90 QRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKGN*ILDGILIANEVVD 149
+R+++Y I+ YK++AK+L+ R++ ++ S++S SQSAF+ G I D +LI ++++
Sbjct: 784 KRVSEYRTIAPCNTQYKIIAKILSKRMQPLLRSIISPSQSAFVPGRAISDNVLITHKILH 605
Query: 150 DARRMDKEL---LLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQKWISECIGTAKAAVL 206
R+ + + + D KAYD + +L V+ + F +W WI EC+ T + L
Sbjct: 604 YLRQSGAKKHVSMAVKTDMTKAYDRIAWNFLREVLTRLGFHGIWISWIMECVSTVSYSFL 425
Query: 207 VNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQLFNGFLVGRVGEVNLT 266
+NG P RGLRQGDPLSP+LF+L E + L +Q G V R +
Sbjct: 424 INGGPQGRVLPSRGLRQGDPLSPYLFILCTEVLSGLCQQALRKGTLPGVKVAR-NCPPIN 248
Query: 267 HLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKSMLT 311
HL FADDT+ G+ + + + +++ + SG +N KS +T
Sbjct: 247 HLLFADDTMFFGKSNASSCAILLSIMDKYRAASGRCIN*TKSAIT 113
>BF650289 weakly similar to GP|9049283|dbj| orf129a {Beta vulgaris}, partial
(69%)
Length = 616
Score = 132 bits (333), Expect = 3e-31
Identities = 64/196 (32%), Positives = 118/196 (59%), Gaps = 1/196 (0%)
Frame = +3
Query: 15 LTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKG 74
L F+ EVK A++ DS K+PG +G + +F K W+++ D + ++DF + G + K
Sbjct: 12 LCSEFTAVEVKNALFSMDSSKAPGIDGYNVHFFKCSWNIIGDSVIDAILDFFKTGFMPKI 191
Query: 75 VNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKG 134
+N T++ L+PK N + ++ PI+ +YK+++K+L +R++ V+ SVVS++QSAF+KG
Sbjct: 192 INCTYVTLLPKEVNVTSVKNFRPIACCSVIYKIISKILTSRMQGVLNSVVSENQSAFVKG 371
Query: 135 N*ILDGILIANEVVDD-ARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQKW 193
I D I++++E+V +R+ + ++D KAYDS + ++ +M+ + FP + W
Sbjct: 372 RVIFDNIILSHELVKSYSRKGISPRCMVKIDLXKAYDSXEWPFIKHLMLELGFPYKFVNW 551
Query: 194 ISECIGTAKAAVLVNG 209
+ + TA NG
Sbjct: 552 VMAXLTTASYTFNXNG 599
>BE248682 similar to GP|18568269|gb putative gag-pol polyprotein {Zea mays},
partial (1%)
Length = 441
Score = 107 bits (268), Expect = 1e-23
Identities = 57/100 (57%), Positives = 71/100 (71%)
Frame = +3
Query: 213 EEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQLFNGFLVGRVGEVNLTHLQFAD 272
EE S++RGL+QGDPL+PFLFLL AEG + LMK LF GF V R G ++HLQ+AD
Sbjct: 6 EEISVQRGLKQGDPLAPFLFLLVAEGISGLMKNAVNRNLFQGFDVKR-GGTRVSHLQYAD 182
Query: 273 DTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKSMLTG 312
DTL IG + N+ +++A+L FE SGLKVNFHKS L G
Sbjct: 183 DTLCIGMPTVDNLWTLKALLQGFEMASGLKVNFHKSSLIG 302
>TC80819 weakly similar to GP|10140689|gb|AAG13524.1 putative non-LTR
retroelement reverse transcriptase {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 1262
Score = 105 bits (263), Expect = 4e-23
Identities = 97/230 (42%), Positives = 114/230 (49%), Gaps = 9/230 (3%)
Frame = +1
Query: 431 GIRFVYQRKKG-GWGLGG*ELLIWHC*ENGVGRC**IKRGCGIEC*KLSMGRLEGGLGKG 489
GIRFV R KG G G LL C V K G K G GG +
Sbjct: 1 GIRFVLLRMKGLGVGAFNLSLLGKWCWRLLVD-----KEGLWHRVLKARYGEEGGGFVRV 165
Query: 490 IDTPQCGGI*FVECVTVQVKELGDGLMRMFVG*LAMGGTLYSGMTLG*GRFRCG*SFLGY 549
+D+ CGG + CV V + G GL + FV *L MGGTL GM G FR *+ L
Sbjct: 166 VDSLLCGGRLYARCVKV*ERGWGTGLKKTFVW*LEMGGTLSFGMIRGQETFR*D*NILAC 345
Query: 550 LSWRWRRRVRWRRCGGWGGAVMEG--SGFG---DVVCLHGR---RRVCCSFM*HCFARHC 601
L WRW R VRW G G A ++G G G VC+ GR V SF * CFA
Sbjct: 346 LIWRWIRSVRW---G*HGEAGVDG*RKGVGVDTPPVCV-GRGVCEGVLYSFK*LCFAG*R 513
Query: 602 S*FLEVDVRPGSRILSESSIPLYYRHRRHG*QVSG**CLV*VYSVKGVFI 651
* +E+ VRP +LSES + ++Y HR + QVSG**CL * YS KG I
Sbjct: 514 Q*QMEMVVRPCEWLLSESLL*IHYFHRSYFRQVSG**CLA*TYSFKGFSI 663
>BG447966 weakly similar to PIR|G96509|G96 protein F27F5.21 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 687
Score = 99.8 bits (247), Expect = 3e-21
Identities = 49/128 (38%), Positives = 74/128 (57%)
Frame = +2
Query: 17 KPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKGVN 76
K F+ EEV +A+ K+PGP+G+ F +++W +V + Q ++ N + +N
Sbjct: 251 KEFTEEEVLEAINQMHPVKAPGPDGLPALFFQKYWHIVGKEVQQMVLQVLNNSMETEELN 430
Query: 77 STFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKGN* 136
TFI LIPK NP DY PISL + K++ KV+ANR++ + V+ QSAF++G
Sbjct: 431 KTFIVLIPKGKNPNTPKDYRPISLCNVVMKIITKVIANRVKQTLPDVIDVEQSAFVQGRL 610
Query: 137 ILDGILIA 144
I D LIA
Sbjct: 611 ITDNALIA 634
>AW736247 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (4%)
Length = 305
Score = 79.3 bits (194), Expect = 4e-15
Identities = 33/58 (56%), Positives = 47/58 (80%), Gaps = 2/58 (3%)
Frame = +1
Query: 19 FSHEEVKQAVWDCDSFKS--PGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKG 74
FS EEV++AVWDCDS S PGP+G++F F+K +W+L+K DF++ +++FH NGKL KG
Sbjct: 121 FSEEEVRKAVWDCDSSHS*NPGPDGVNFTFVKEYWELIKVDFLRVVMEFHTNGKLGKG 294
>TC90516
Length = 983
Score = 77.8 bits (190), Expect = 1e-14
Identities = 43/134 (32%), Positives = 75/134 (55%)
Frame = -2
Query: 15 LTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKG 74
L+ F +++ V D +SP P+G + NF R +++K D F+ L+K
Sbjct: 532 LSAQFLVSKMELVVSSLDGNESPRPDGFNLNFFIRLRNMLKADIEIMFEQFYTPANLLKI 353
Query: 75 VNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKG 134
+ F+ LI KV P L D+ +S +G L K++AKVLA RL +++ ++ +QS F++G
Sbjct: 352 FS*YFLTLISKVEYPILLGDFSLMSFLGSL*KLMAKVLALRLAHIMEKIIFVNQSTFVRG 173
Query: 135 N*ILDGILIANEVV 148
+DG++ NE++
Sbjct: 172 RQHVDGVVAINEII 131
>BG587108 similar to PIR|G96509|G9 protein F27F5.21 [imported] - Arabidopsis
thaliana, partial (17%)
Length = 677
Score = 73.9 bits (180), Expect(2) = 3e-14
Identities = 35/90 (38%), Positives = 57/90 (62%)
Frame = +3
Query: 45 NFIKRFWDLVKDDFMQFLVDFHRNGKLIKGVNSTFIALIPKVNNPQRLNDYMPISLVGCL 104
+F + W ++K D ++ + F +GKL +N+T I LIPK P R+ + PISL
Sbjct: 408 SFFQHSWHIIKMDLLKMVNSFLASGKLDTRLNTTNICLIPKKKRPTRMTELRPISLCNVG 587
Query: 105 YKVLAKVLANRLRNVIGSVVSDSQSAFIKG 134
YK+++KVL RL+ + S++S++QSAF+ G
Sbjct: 588 YKIISKVLCQRLKVCLPSLISETQSAFVHG 677
Score = 22.7 bits (47), Expect(2) = 3e-14
Identities = 9/24 (37%), Positives = 17/24 (70%)
Frame = +2
Query: 20 SHEEVKQAVWDCDSFKSPGPNGIS 43
+ EEV+ A++ K+PGP+G++
Sbjct: 332 TEEEVRLALFIMHPEKAPGPDGMT 403
>BG647708 weakly similar to GP|13786450|gb| putative reverse transcriptase
{Oryza sativa}, partial (9%)
Length = 708
Score = 71.6 bits (174), Expect = 9e-13
Identities = 38/91 (41%), Positives = 54/91 (58%)
Frame = +1
Query: 218 ERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQLFNGFLVGRVGEVNLTHLQFADDTLII 277
E+GLRQGDPLSP+LF+L A + L+K+ Q +G V R + +THL FADD+L+
Sbjct: 7 EKGLRQGDPLSPYLFILCANVLSGLLKREGNKQNLHGIQVAR-SDPKITHLLFADDSLLF 183
Query: 278 GEKSWLNVRSMRAVLMLFEEVSGLKVNFHKS 308
+ ++ VL ++ SG VNF KS
Sbjct: 184 ARANLTEAATIMQVLHSYQSASGQLVNFEKS 276
>BG586267 weakly similar to PIR|A84888|A8 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana, partial (16%)
Length = 794
Score = 61.6 bits (148), Expect = 9e-10
Identities = 32/89 (35%), Positives = 47/89 (51%)
Frame = +3
Query: 15 LTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKG 74
L S EEV++AV+D + K PGP+G++ F ++FWD + DD +F R GKL +G
Sbjct: 399 LMAQISREEVREAVFDINPHKCPGPDGMNVFFFQQFWDTMGDDLTSMAQEFLRTGKLEEG 578
Query: 75 VNSTFIALIPKVNNPQRLNDYMPISLVGC 103
+N T I K + + P V C
Sbjct: 579 INKTNIWFGSKKAGGKEASGISPHKFV*C 665
Score = 55.5 bits (132), Expect = 6e-08
Identities = 32/103 (31%), Positives = 58/103 (56%)
Frame = +1
Query: 43 SFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKGVNSTFIALIPKVNNPQRLNDYMPISLVG 102
SFN W+++ + + + G L + L+PK +RL ++ PISL
Sbjct: 493 SFNNFGTQWEMISHLWRK---NSSEQGNLKRESTKQTSGLVPKKLEAKRLVEFRPISLCN 663
Query: 103 CLYKVLAKVLANRLRNVIGSVVSDSQSAFIKGN*ILDGILIAN 145
YK+++KVL+ RL++V+ +++++Q+AF + I D ILIA+
Sbjct: 664 VAYKIVSKVLSKRLKSVLPWIITETQAAFGRRQLISDNILIAH 792
>AW774658 similar to GP|2808681|emb| Hcr9-4B {Lycopersicon hirsutum}, partial
(4%)
Length = 665
Score = 36.2 bits (82), Expect(2) = 7e-06
Identities = 14/19 (73%), Positives = 17/19 (88%)
Frame = -1
Query: 266 THLQFADDTLIIGEKSWLN 284
+HLQFADDTL++G KSW N
Sbjct: 383 SHLQFADDTLLLGVKSWAN 327
Score = 32.0 bits (71), Expect(2) = 7e-06
Identities = 12/28 (42%), Positives = 22/28 (77%)
Frame = -3
Query: 283 LNVRSMRAVLMLFEEVSGLKVNFHKSML 310
L R++R++L++FE +SGLKVN + ++
Sbjct: 336 LGERALRSILVIFENMSGLKVNLREEVI 253
>TC91765 weakly similar to GP|19881779|gb|AAM01180.1 Putative retroelement
{Oryza sativa (japonica cultivar-group)}, partial (1%)
Length = 625
Score = 40.0 bits (92), Expect = 0.003
Identities = 18/47 (38%), Positives = 29/47 (61%)
Frame = +2
Query: 197 CIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLM 243
C+ + VLVN ++ RGL+QGD LSP++F++ EG + L+
Sbjct: 104 CVESNDYYVLVNNDAVDPIIPSRGLQQGDHLSPYIFIICVEGLSFLI 244
>TC82520
Length = 833
Score = 31.6 bits (70), Expect(2) = 0.47
Identities = 19/34 (55%), Positives = 20/34 (57%), Gaps = 2/34 (5%)
Frame = +1
Query: 579 VVCLHGRR--RVCCSFM*HCFARHCS*FLEVDVR 610
VVCL G V C *HCFAR CS* L + VR
Sbjct: 1 VVCLGGGECEGVVCFTS*HCFARKCS*CLPLAVR 102
Score = 19.6 bits (39), Expect(2) = 0.47
Identities = 9/17 (52%), Positives = 11/17 (63%)
Frame = +2
Query: 635 SG**CLV*VYSVKGVFI 651
S * CL YS KGV++
Sbjct: 173 SS*RCLAKEYSFKGVYV 223
>BG582075 similar to PIR|T05242|T052 hypothetical protein F18A5.120 -
Arabidopsis thaliana, partial (21%)
Length = 784
Score = 32.0 bits (71), Expect = 0.77
Identities = 28/75 (37%), Positives = 36/75 (47%), Gaps = 9/75 (12%)
Frame = -2
Query: 510 ELGDGLMRMFVG*LAMGGTLYSGMTLG*GRFRCG*SFLGYLS---------WRWRRRVRW 560
+LG GL + G + GG+ G GR R G*S G ++ RWRRR RW
Sbjct: 309 QLGLGLSQTGDGHVIGGGSRSGG-----GRRRGG*SSNGIVN*GRIFYDGDRRWRRRFRW 145
Query: 561 RRCGGWGGAVMEGSG 575
R G G A++ G G
Sbjct: 144 VRSG--GEAILFGGG 106
>AJ503675 similar to GP|21928168|gb| AT5g23040/MYJ24_3 {Arabidopsis
thaliana}, partial (20%)
Length = 576
Score = 30.8 bits (68), Expect = 1.7
Identities = 15/30 (50%), Positives = 18/30 (60%)
Frame = -2
Query: 371 F*CLTGSSEVCLVLSFGLLSFLFQSPCRYN 400
F*C G + C LS GLLSFL PC ++
Sbjct: 506 F*CRFGLQDFCHQLS*GLLSFLGSRPCHWS 417
>TC77945 similar to SP|O24296|GSHY_PEA Phospholipid hydroperoxide
glutathione peroxidase chloroplast precursor (EC
1.11.1.9) (PHGPx)., complete
Length = 1006
Score = 30.4 bits (67), Expect = 2.2
Identities = 26/75 (34%), Positives = 41/75 (54%), Gaps = 16/75 (21%)
Frame = +1
Query: 15 LTKPFSHEEVKQAVWDCDSFKSPGP-------NGIS----FNFIKR-----FWDLVKDDF 58
+ +P S+EE+K+ + C FK+ P NG + F+K F DLVK +F
Sbjct: 520 MQEPGSNEEIKK--FACTRFKAEFPIFDKVDVNGPFTAPVYQFLKSSSGGFFGDLVKWNF 693
Query: 59 MQFLVDFHRNGKLIK 73
+FLVD +NGK+++
Sbjct: 694 EKFLVD--KNGKVVE 732
>BQ144057 similar to GP|1483567|emb| viral proteinase {Pseudorabies virus},
partial (5%)
Length = 1295
Score = 30.4 bits (67), Expect = 2.2
Identities = 19/47 (40%), Positives = 25/47 (52%)
Frame = -1
Query: 443 WGLGG*ELLIWHC*ENGVGRC**IKRGCGIEC*KLSMGRLEGGLGKG 489
W LG LL+W G+G C ++ CG+ C L +GR G LG G
Sbjct: 347 WVLGFDVLLVWWV-LYGMGWCVCVEENCGVRCGILRVGR-AGWLGAG 213
>TC83401 homologue to GP|10441918|gb|AAG17236.1 unknown {Homo sapiens},
partial (3%)
Length = 374
Score = 29.6 bits (65), Expect = 3.8
Identities = 18/42 (42%), Positives = 23/42 (53%), Gaps = 2/42 (4%)
Frame = +2
Query: 379 EVCLVLSFGLLSFLFQSPCRYNFLY*IYF--KKKKKLGGGGG 418
EVC + F F F P ++ ++Y Y KKKKKL GG G
Sbjct: 212 EVCPINLF----FYF*EPIKFIYIYKXYLEXKKKKKLEGGAG 325
>TC86434 similar to GP|2262100|gb|AAB63608.1| unknown protein {Arabidopsis
thaliana}, partial (83%)
Length = 1480
Score = 29.3 bits (64), Expect = 5.0
Identities = 20/66 (30%), Positives = 32/66 (48%)
Frame = +3
Query: 66 HRNGKLIKGVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVS 125
H G++S L+P +++P R N + P+S +G L +L VL + GSV
Sbjct: 141 HLRNLKASGLHSVGFPLLPIISSPLR-NAFFPLSRLGMLKSMLI*VLVLLALKLDGSVQP 317
Query: 126 DSQSAF 131
+ AF
Sbjct: 318 VTSHAF 335
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.344 0.156 0.540
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,967,219
Number of Sequences: 36976
Number of extensions: 378703
Number of successful extensions: 4516
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 2520
Number of HSP's successfully gapped in prelim test: 211
Number of HSP's that attempted gapping in prelim test: 1729
Number of HSP's gapped (non-prelim): 3023
length of query: 651
length of database: 9,014,727
effective HSP length: 102
effective length of query: 549
effective length of database: 5,243,175
effective search space: 2878503075
effective search space used: 2878503075
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147000.3


