

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147000.2 - phase: 0
(840 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
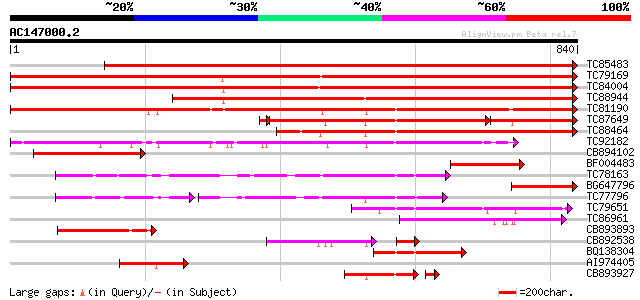
Score E
Sequences producing significant alignments: (bits) Value
TC85483 type IIB calcium ATPase MCA5 [Medicago truncatula] 1199 0.0
TC79169 type IIB calcium ATPase [Medicago truncatula] 1132 0.0
TC84004 type IIB calcium ATPase [Medicago truncatula] 1109 0.0
TC88944 type IIB calcium ATPase [Medicago truncatula] 930 0.0
TC81190 type IIB calcium ATPase [Medicago truncatula] 770 0.0
TC87649 similar to SP|Q9SZR1|ACAA_ARATH Potential calcium-transp... 358 e-128
TC88464 similar to SP|Q9LU41|ACA9_ARATH Potential calcium-transp... 451 e-127
TC92182 type IIA calcium ATPase [Medicago truncatula] 339 2e-93
CB894102 similar to GP|8843813|db Ca2+-transporting ATPase-like ... 191 8e-49
BF004483 GP|21314227| type IIB calcium ATPase MCA5 {Medicago tru... 181 8e-46
TC78163 H+-ATPase 173 2e-43
BG647796 GP|21314227|g type IIB calcium ATPase MCA5 {Medicago tr... 166 3e-41
TC77796 H+-ATPase 117 2e-40
TC79651 similar to PIR|T52581|T52581 Ca2+-transporting ATPase (E... 136 3e-32
TC86961 similar to GP|1742951|emb|CAA70946.1 Ca2+-ATPase {Arabid... 130 2e-30
CB893893 similar to GP|8843813|db Ca2+-transporting ATPase-like ... 126 3e-29
CB892538 similar to SP|Q9XES1|ECA Calcium-transporting ATPase 4 ... 91 1e-28
BQ138304 homologue to GP|6688833|emb putative calcium P-type ATP... 113 3e-25
AI974405 similar to SP|Q9LF79|ACA Calcium-transporting ATPase 8 ... 94 3e-19
CB893927 homologue to GP|17342714|g type IIA calcium ATPase {Med... 83 1e-18
>TC85483 type IIB calcium ATPase MCA5 [Medicago truncatula]
Length = 2541
Score = 1199 bits (3103), Expect = 0.0
Identities = 601/702 (85%), Positives = 652/702 (92%), Gaps = 2/702 (0%)
Frame = +2
Query: 141 GSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATIIGKIGLVFAVITFTVLV 200
GSC MLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATIIGKIGL FA++TF VLV
Sbjct: 2 GSCKMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATIIGKIGLFFAIVTFAVLV 181
Query: 201 KGHLSHKIREGNFWRWTGDNAMEMLEYFAIAVTIVVVAVPEGLPLAVTLSLAFAMKKMMN 260
+G +S K+++ NFW W GD+A+EMLEYFAIAVTIVVVAVPEGLPLAVTLSLAFAMKKMMN
Sbjct: 182 QGLVSLKLQQENFWNWNGDDALEMLEYFAIAVTIVVVAVPEGLPLAVTLSLAFAMKKMMN 361
Query: 261 DKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCICMNSKEVSNSSSS--SDIPD 318
DKALVR+LAACETMGSATTICSDKTGTLTTN MTVVKTCICM SKEVSN +SS S++P+
Sbjct: 362 DKALVRNLAACETMGSATTICSDKTGTLTTNHMTVVKTCICMKSKEVSNKTSSLCSELPE 541
Query: 319 SAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTETAILEFGLSLGGDSKAEREACKIVKV 378
S KLL QSIFNNTGGEVV NK+GK EILGTPTETAILEFGLSLGGD + ER+ACK+VKV
Sbjct: 542 SVVKLLQQSIFNNTGGEVVVNKQGKHEILGTPTETAILEFGLSLGGDFQGERQACKLVKV 721
Query: 379 EPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAACDKVIDLNGDVVALDGESTNYLNSI 438
EPFNS KKRMG VVE P G +RAHCKGASEI+LAACDKV++ NG+VV LD ESTN+L +
Sbjct: 722 EPFNSTKKRMGAVVELPSGGLRAHCKGASEIVLAACDKVLNSNGEVVPLDEESTNHLTNT 901
Query: 439 INQFANEALRTLCLAYMELENGFAAEDPIPASGYTCIGIVGIKDPVRPGVKQSVAECRSA 498
INQFANEALRTLCLAYMELENGF+AED IP +GYTCIG+VGIKDPVRPGVK+SVA CRSA
Sbjct: 902 INQFANEALRTLCLAYMELENGFSAEDTIPVTGYTCIGVVGIKDPVRPGVKESVALCRSA 1081
Query: 499 GIVVRMVTGDNINTAKAIARECGILTDDGIAIEGPDFREKTQEELFELIPKIQVMARSSP 558
GI VRMVTGDNINTAKAIARECGILTDDGIAIEGP+FREK+ EEL ELIPKIQVMARSSP
Sbjct: 1082GITVRMVTGDNINTAKAIARECGILTDDGIAIEGPEFREKSLEELLELIPKIQVMARSSP 1261
Query: 559 LDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDN 618
LDKHTLV+ LRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDN
Sbjct: 1262LDKHTLVRHLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDN 1441
Query: 619 FSTIVTVARWGRSVYINIQKFVQFQLTVNVVALLVNFTSACMTGSAPLTAVQLLWVNMIM 678
FSTIVTVA+WGRSVYINIQKFVQFQLTVN+VAL+VNFTSAC+TG+APLTAVQLLWVNMIM
Sbjct: 1442FSTIVTVAKWGRSVYINIQKFVQFQLTVNIVALIVNFTSACLTGTAPLTAVQLLWVNMIM 1621
Query: 679 DTLGALALATEPPTDDLMKREPVGRKGDFINNVMWRNILGQALYQFVVIWFLQSVGKWVF 738
DTLGALALATEPP DDLMKR PVGRKG+FI+NVMWRNILGQ+LYQF+VIWFLQS GK +F
Sbjct: 1622DTLGALALATEPPNDDLMKRAPVGRKGNFISNVMWRNILGQSLYQFMVIWFLQSKGKTIF 1801
Query: 739 FLRGPNADIVLNTLIFNTFVFCQVFNEINSREMEEIDVFKGIWDNHVFVAVISATVVFQI 798
L GPN+D+VLNTLIFN FVFCQVFNEINSREME+I+VFKGI DN+VFV VISAT+ FQI
Sbjct: 1802SLDGPNSDLVLNTLIFNAFVFCQVFNEINSREMEKINVFKGILDNYVFVGVISATIFFQI 1981
Query: 799 IIVEYLGTFANTTPLSLVQWIFCLGVGYMGMPIAVRLKQIPV 840
IIVEYLGTFANTTPL+LVQW FCL VG+MGMPIA RLK+IPV
Sbjct: 1982IIVEYLGTFANTTPLTLVQWFFCLFVGFMGMPIAARLKKIPV 2107
>TC79169 type IIB calcium ATPase [Medicago truncatula]
Length = 3565
Score = 1132 bits (2927), Expect = 0.0
Identities = 567/844 (67%), Positives = 693/844 (81%), Gaps = 4/844 (0%)
Frame = +2
Query: 1 MTLMILGVCAFVSLIVGVLTEGWPKGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLDK 60
+TL+IL VCA VS+ +G+ TEGWPKG +DG+GI+ SI LVV VTA SDY+QSLQF DLDK
Sbjct: 677 LTLIILIVCALVSIGIGLPTEGWPKGVYDGVGILLSIFLVVTVTAVSDYQQSLQFLDLDK 856
Query: 61 EKKKISIQVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGES 120
EKKKISI VTR+G RQK+SIY+L+ GDIVHL+ GDQVPADG+F+ G+S+LIDESSL+GES
Sbjct: 857 EKKKISIHVTRDGKRQKVSIYDLVVGDIVHLSTGDQVPADGIFIQGYSLLIDESSLSGES 1036
Query: 121 EPIMVTTQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGV 180
EP+ + + PFLLSGTKVQDG M+VTTVGMRT+WGKLM TLSEGG+DETPLQVKLNGV
Sbjct: 1037 EPVDIDNRRPFLLSGTKVQDGQAKMIVTTVGMRTEWGKLMETLSEGGEDETPLQVKLNGV 1216
Query: 181 ATIIGKIGLVFAVITFTVLVKGHLSHKIREGNFWRWTGDNAMEMLEYFAIAVTIVVVAVP 240
AT+IGKIGL FAV+TF VL + K G+F W+ ++A+++L+YFAIAVTI+VVA+P
Sbjct: 1217 ATVIGKIGLTFAVLTFLVLTARFVIEKAINGDFTSWSSEDALKLLDYFAIAVTIIVVAIP 1396
Query: 241 EGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCI 300
EGLPLAVTLSLAFAMKK+MND+ALVRHL+ACETMGSA+ IC+DKTGTLTTN M V K I
Sbjct: 1397 EGLPLAVTLSLAFAMKKLMNDRALVRHLSACETMGSASCICTDKTGTLTTNHMVVDKIWI 1576
Query: 301 CMNSKEVSNSSSS----SDIPDSAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTETAIL 356
C + E+ S+ S+I D +LLQ+IF NT EVV + +GK+ ILGTPTE+A+L
Sbjct: 1577 CEKTVEMKGDESTDKLKSEISDEVLSILLQAIFQNTSSEVVKDNEGKQTILGTPTESALL 1756
Query: 357 EFGLSLGGDSKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAACDK 416
EFGL GGD A+R +CK++KVEPFNS++K+M V+V PDG VRA CKGASEI+L CDK
Sbjct: 1757 EFGLVSGGDFDAQRRSCKVLKVEPFNSDRKKMSVLVGLPDGGVRAFCKGASEIVLKMCDK 1936
Query: 417 VIDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDPIPASGYTCIG 476
+ID NG + L E ++ II+ FANEALRTLCLA +++ E IP +GYT I
Sbjct: 1937 IIDSNGTTIDLPEEKARIVSDIIDGFANEALRTLCLAVKDIDET-QGETNIPENGYTLIT 2113
Query: 477 IVGIKDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTDDGIAIEGPDFR 536
IVGIKDPVRPGVK++V +C +AGI VRMVTGDNINTAKAIA+ECGILT+ G+AIEGP+FR
Sbjct: 2114 IVGIKDPVRPGVKEAVQKCLAAGISVRMVTGDNINTAKAIAKECGILTEGGVAIEGPEFR 2293
Query: 537 EKTQEELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGL 596
++E++ ++IP+IQVMARS PLDKHTLV +LR FGEVVAVTGDGTNDAPALHE+DIGL
Sbjct: 2294 NLSEEQMKDIIPRIQVMARSLPLDKHTLVTRLRNMFGEVVAVTGDGTNDAPALHESDIGL 2473
Query: 597 AMGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNVVALLVNFT 656
AMGIAGTEVAKE+ADVII+DDNF+TIV VA+WGR++YINIQKFVQFQLTVNVVAL+ NF
Sbjct: 2474 AMGIAGTEVAKENADVIIMDDNFTTIVKVAKWGRAIYINIQKFVQFQLTVNVVALITNFV 2653
Query: 657 SACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKREPVGRKGDFINNVMWRNI 716
SAC+TG+APLTAVQLLWVN+IMDTLGALALATEPP D LM+R+PVGRK FI MWRNI
Sbjct: 2654 SACITGAAPLTAVQLLWVNLIMDTLGALALATEPPNDGLMERQPVGRKASFITKPMWRNI 2833
Query: 717 LGQALYQFVVIWFLQSVGKWVFFLRGPNADIVLNTLIFNTFVFCQVFNEINSREMEEIDV 776
GQ+LYQ +V+ L GK + L GP++ VLNTLIFN+FVFCQVFNEINSRE+E+I++
Sbjct: 2834 FGQSLYQLIVLGVLNFEGKRLLGLSGPDSTAVLNTLIFNSFVFCQVFNEINSREIEKINI 3013
Query: 777 FKGIWDNHVFVAVISATVVFQIIIVEYLGTFANTTPLSLVQWIFCLGVGYMGMPIAVRLK 836
F+G++D+ +F++VI AT VFQ+IIVE+LGTFA+T PL+ W+ L G + MP+A LK
Sbjct: 3014 FRGMFDSWIFLSVILATAVFQVIIVEFLGTFASTVPLTWQFWLLSLLFGVLSMPLAAILK 3193
Query: 837 QIPV 840
IPV
Sbjct: 3194 CIPV 3205
>TC84004 type IIB calcium ATPase [Medicago truncatula]
Length = 3655
Score = 1109 bits (2869), Expect = 0.0
Identities = 557/846 (65%), Positives = 685/846 (80%), Gaps = 6/846 (0%)
Frame = +1
Query: 1 MTLMILGVCAFVSLIVGVLTEGWPKGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLDK 60
+TL IL VCA VS+ +G+ TEGWPKG +DG+GI+ SI LVV VTA SDYRQSLQF DLD+
Sbjct: 745 LTLTILMVCAVVSIGIGLATEGWPKGTYDGVGIILSIFLVVIVTAVSDYRQSLQFMDLDR 924
Query: 61 EKKKISIQVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGES 120
EKKKI +QV R+G R+K+SIY+++ GDI+HL+ GDQVPADG+++SG+S+LIDESSL+GES
Sbjct: 925 EKKKIFVQVNRDGKRKKISIYDVVVGDIIHLSTGDQVPADGIYISGYSLLIDESSLSGES 1104
Query: 121 EPIMVTTQNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGV 180
EP+ +T ++PFLLSGTKVQDG MLVTTVGMRT+WGKLM TL+EGG+DETPLQVKLNGV
Sbjct: 1105 EPVFITEEHPFLLSGTKVQDGQGKMLVTTVGMRTEWGKLMETLNEGGEDETPLQVKLNGV 1284
Query: 181 ATIIGKIGLVFAVITFTVLVKGHLSHKIREGNFWRWTGDNAMEMLEYFAIAVTIVVVAVP 240
ATIIGKIGL FA++TF VL L K G F W+ ++A ++L++FAIAVTI+VVAVP
Sbjct: 1285 ATIIGKIGLFFAIVTFLVLTVRFLVEKALHGEFGNWSSNDATKLLDFFAIAVTIIVVAVP 1464
Query: 241 EGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCI 300
EGLPLAVTLSLAFAMKK+MND ALVRHL+ACETMGSA+ IC+DKTGTLTTN M V K I
Sbjct: 1465 EGLPLAVTLSLAFAMKKLMNDMALVRHLSACETMGSASCICTDKTGTLTTNHMVVNKIWI 1644
Query: 301 CMNSKEVSNSSSSSD----IPDSAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTETAIL 356
C N+ ++ S+ + I + +LLQ+IF NT EVV +K GK ILG+PTE+A+L
Sbjct: 1645 CENTTQLKGDESADELKTNISEGVLSILLQAIFQNTSAEVVKDKNGKNTILGSPTESALL 1824
Query: 357 EFGLSLGG--DSKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAAC 414
EFGL LG D++ +A KI+K+EPFNS +K+M V+V P+G V+A CKGASEIIL C
Sbjct: 1825 EFGLLLGSEFDARNHSKAYKILKLEPFNSVRKKMSVLVGLPNGRVQAFCKGASEIILEMC 2004
Query: 415 DKVIDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDPIPASGYTC 474
DK+ID NG+VV L + N ++ +IN FA+EALRTLCLA ++ N E IP SGYT
Sbjct: 2005 DKMIDCNGEVVDLPADRANIVSDVINSFASEALRTLCLAVRDI-NETQGETNIPDSGYTL 2181
Query: 475 IGIVGIKDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTDDGIAIEGPD 534
I +VGIKDPVRPGVK++V C +AGI VRMVTGDNINTAKAIA+ECGILTDDG+AIEGP
Sbjct: 2182 IALVGIKDPVRPGVKEAVQTCIAAGITVRMVTGDNINTAKAIAKECGILTDDGVAIEGPS 2361
Query: 535 FREKTQEELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADI 594
FRE + E++ ++IP+IQVMARS PLDKH LV LR FGEVVAVTGDGTNDAPALHEADI
Sbjct: 2362 FRELSDEQMKDIIPRIQVMARSLPLDKHKLVTNLRNMFGEVVAVTGDGTNDAPALHEADI 2541
Query: 595 GLAMGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNVVALLVN 654
GLAMGIAGTEVAKE ADVII+DDNF+TIV V +WGR+VYINIQKFVQFQLTVNVVAL++N
Sbjct: 2542 GLAMGIAGTEVAKEKADVIIMDDNFATIVNVVKWGRAVYINIQKFVQFQLTVNVVALIIN 2721
Query: 655 FTSACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKREPVGRKGDFINNVMWR 714
F SAC+TGSAPLTAVQLLWVN+IMDTLGALALATEPP D L+KR PVGR FI MWR
Sbjct: 2722 FVSACITGSAPLTAVQLLWVNLIMDTLGALALATEPPNDGLLKRPPVGRGASFITKTMWR 2901
Query: 715 NILGQALYQFVVIWFLQSVGKWVFFLRGPNADIVLNTLIFNTFVFCQVFNEINSREMEEI 774
NI+GQ++YQ +V+ L GK + + G +A VLNTLIFN+FVFCQVFNEINSR++E+I
Sbjct: 2902 NIIGQSIYQLIVLAILNFDGKRLLGINGSDATEVLNTLIFNSFVFCQVFNEINSRDIEKI 3081
Query: 775 DVFKGIWDNHVFVAVISATVVFQIIIVEYLGTFANTTPLSLVQWIFCLGVGYMGMPIAVR 834
++F+G++D+ +F+ +I +TV FQ++IVE+LG FA+T PLS W+ + +G + MP+AV
Sbjct: 3082 NIFRGMFDSWIFLLIIFSTVAFQVVIVEFLGAFASTVPLSWQLWLLSVLIGAISMPLAVI 3261
Query: 835 LKQIPV 840
+K IPV
Sbjct: 3262 VKCIPV 3279
>TC88944 type IIB calcium ATPase [Medicago truncatula]
Length = 1883
Score = 930 bits (2403), Expect = 0.0
Identities = 467/604 (77%), Positives = 531/604 (87%), Gaps = 4/604 (0%)
Frame = +3
Query: 241 EGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCI 300
+GLPLAVTL LAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTN MTVVK CI
Sbjct: 3 DGLPLAVTLILAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNHMTVVKACI 182
Query: 301 CMNSKEVSNSSSSSD----IPDSAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTETAIL 356
C KEV++S SSD +PDSA +LL+SIFNNTGGEVV N+ GK EILG+PTETAIL
Sbjct: 183 CGKIKEVNSSIDSSDFSSDLPDSAIAILLESIFNNTGGEVVKNENGKIEILGSPTETAIL 362
Query: 357 EFGLSLGGDSKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAACDK 416
EFGLSLGGD ER+A K+VKVEPFNS KKRMGVV++ PDG RAHCKGASEIILAACDK
Sbjct: 363 EFGLSLGGDFHKERQASKLVKVEPFNSIKKRMGVVLQLPDGGYRAHCKGASEIILAACDK 542
Query: 417 VIDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDPIPASGYTCIG 476
+D N +V LD +S ++LN I +FANEALRTLCLAY+++ + F PIP +GYTC+G
Sbjct: 543 FVDSNSKIVPLDEDSISHLNDTIEKFANEALRTLCLAYIDIHDEFLVGSPIPVNGYTCVG 722
Query: 477 IVGIKDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTDDGIAIEGPDFR 536
IVGIKDPVRPGV++SVA CRSAGI VRMVTGDNINTAKAIARECGILTD GIAIEGP+FR
Sbjct: 723 IVGIKDPVRPGVRESVAICRSAGITVRMVTGDNINTAKAIARECGILTD-GIAIEGPEFR 899
Query: 537 EKTQEELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGL 596
E +++EL ++IPKIQVMARSSP+DKHTLVK LRTTF EVVAVTGDGTNDAPALHEADIGL
Sbjct: 900 EMSEKELLDIIPKIQVMARSSPMDKHTLVKHLRTTFEEVVAVTGDGTNDAPALHEADIGL 1079
Query: 597 AMGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNVVALLVNFT 656
AMGIAGTEVAKESADVIILDDNFSTIVTVA+WGRSVYINIQKFVQFQLTVNVVAL+VNFT
Sbjct: 1080AMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVALIVNFT 1259
Query: 657 SACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKREPVGRKGDFINNVMWRNI 716
SAC+TG+APLTAVQLLWVNMIMDTLGALALATEPP D+LMKR PVGRKG+FI+NVMWRNI
Sbjct: 1260SACLTGNAPLTAVQLLWVNMIMDTLGALALATEPPNDELMKRAPVGRKGNFISNVMWRNI 1439
Query: 717 LGQALYQFVVIWFLQSVGKWVFFLRGPNADIVLNTLIFNTFVFCQVFNEINSREMEEIDV 776
GQ++YQFV+IW LQ+ GK VF L GP++D++LNTLIFN+FVFCQVFNEI+SR+ME I+V
Sbjct: 1440TGQSIYQFVIIWLLQTRGKTVFHLDGPDSDLILNTLIFNSFVFCQVFNEISSRDMERINV 1619
Query: 777 FKGIWDNHVFVAVISATVVFQIIIVEYLGTFANTTPLSLVQWIFCLGVGYMGMPIAVRLK 836
F+GI N+VF AV++ T +FQIIIVE+LGT+ANT+PLSL W+ + +G +GMPI LK
Sbjct: 1620FEGILKNYVFTAVLTCTAIFQIIIVEFLGTYANTSPLSLKLWLISVFLGVLGMPIGAALK 1799
Query: 837 QIPV 840
IPV
Sbjct: 1800MIPV 1811
>TC81190 type IIB calcium ATPase [Medicago truncatula]
Length = 3277
Score = 770 bits (1989), Expect = 0.0
Identities = 429/867 (49%), Positives = 576/867 (65%), Gaps = 28/867 (3%)
Frame = +1
Query: 2 TLMILGVCAFVSLIVGVLTEGWPKGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLDKE 61
T++IL VCA +SL G+ G +G ++G I ++ LVV V+A S++RQ QF L K
Sbjct: 541 TIIILLVCAGLSLGFGIKEHGPGEGWYEGGSIFLAVFLVVVVSALSNFRQERQFHKLSKI 720
Query: 62 KKKISIQVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGESE 121
I ++V RNG Q++SI+++L GDIV L IGDQ+PADG+F+SG+S+ +DESS+TGES+
Sbjct: 721 SNNIKVEVVRNGRPQQISIFDVLVGDIVSLKIGDQIPADGVFLSGYSLQVDESSMTGESD 900
Query: 122 PIMVTT-QNPFLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGV 180
+ + + PFLLSG KV DG MLVT+VG T WG++M+++S ++ TPLQ +L+ +
Sbjct: 901 HVEIEPLRAPFLLSGAKVVDGYAQMLVTSVGKNTSWGQMMSSISRDTNERTPLQARLDKL 1080
Query: 181 ATIIGKIGLVFAVITFTVLVKGHL---SHKIREGNFWRWT----GDNAMEMLEYFAIAVT 233
+ IGK+GL A + VL+ + SH + +R + D ++ A AVT
Sbjct: 1081 TSSIGKVGLAVAFLVLLVLLIRYFTGNSHDEKGNKEFRGSKTDINDVMNSVVSIVAAAVT 1260
Query: 234 IVVVAVPEGLPLAVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRM 293
IVVVA+PEGLPLAVTL+LA++MK+MM D A+VR L+ACETMGSAT IC+DKTGTLT N+M
Sbjct: 1261 IVVVAIPEGLPLAVTLTLAYSMKRMMADHAMVRKLSACETMGSATVICTDKTGTLTLNQM 1440
Query: 294 TVVKTCICMNSKEVSNSSSSSDIPDSAAKLLLQSIFNNTGGEVVYNKKGKR-EILGTPTE 352
V K C+ + + + S++ P +L Q + NT G V G EI G+PTE
Sbjct: 1441 RVTK--FCLGPENIIENFSNAMTP-KVLELFHQGVGLNTTGSVYNPPSGSEPEISGSPTE 1611
Query: 353 TAILEFG-LSLGGDSKAEREACKIVKVEPFNSEKKRMGVVV--EQPDGSVRAHCKGASEI 409
AIL + L LG D ++ K++ VE FNSEKKR GV + E D SV H KGA+E+
Sbjct: 1612 KAILMWAVLDLGMDMDEMKQKHKVLHVETFNSEKKRSGVAIRKENDDNSVHVHWKGAAEM 1791
Query: 410 ILAACDKVIDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELENGF-------- 461
ILA C ID NG +LD E + + II A +LR + A+ E+ +
Sbjct: 1792 ILAMCTNYIDSNGARKSLDEEERSKIERIIQVMAASSLRCIAFAHTEISDSEDIDYMIKR 1971
Query: 462 --AAEDPIPASGYTCIGIVGIKDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARE 519
+ + G T +GIVG+KDP RP K++V C++AG+ ++M+TGDNI TAKAIA E
Sbjct: 1972 EKKSHQMLREDGLTLLGIVGLKDPCRPNTKKAVETCKAAGVEIKMITGDNIFTAKAIAIE 2151
Query: 520 CGILTDD------GIAIEGPDFREKTQEELFELIPKIQVMARSSPLDKHTLVKQLRTTFG 573
CGIL + G +EG +FR T+EE E + I+VMARSSP+DK +V+ LR G
Sbjct: 2152 CGILDSNSDHAKAGEVVEGVEFRSYTEEERMEKVDNIRVMARSSPMDKLLMVQCLRKK-G 2328
Query: 574 EVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVY 633
VVAVTGDGTNDAPAL EADIGL+MGI GTEVAKES+D++ILDDNF+++ TV RWGR VY
Sbjct: 2329 HVVAVTGDGTNDAPALKEADIGLSMGIQGTEVAKESSDIVILDDNFNSVATVLRWGRCVY 2508
Query: 634 INIQKFVQFQLTVNVVALLVNFTSACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTD 693
NIQKF+QFQLTVNV AL++NF +A +G PLT VQLLWVN+IMDTLGALALATE PT
Sbjct: 2509 NNIQKFIQFQLTVNVAALVINFIAAVSSGDVPLTTVQLLWVNLIMDTLGALALATERPTK 2688
Query: 694 DLMKREPVGRKGDFINNVMWRNILGQALYQFVVIWFLQSVGKWVFFLRGPNADIVLNTLI 753
+LMK++P+GR I N+MWRN+L QA YQ V+ +Q GK +F + + V +TLI
Sbjct: 2689 ELMKKKPIGRTAPLITNIMWRNLLAQASYQIAVLLIMQFYGKSIFNV----SKEVKDTLI 2856
Query: 754 FNTFVFCQVFNEINSREMEEIDVFKGIWDNHVFVAVISATVVFQIIIVEYLGTFANTTPL 813
FNTFV CQVFNE NSR ME++ VF+GI NH+F+ +I T+V QI++VE L FA+T L
Sbjct: 2857 FNTFVLCQVFNEFNSRSMEKLYVFEGILKNHLFLGIIGITIVLQILMVELLRKFADTERL 3036
Query: 814 SLVQWIFCLGVGYMGMPIAVRLKQIPV 840
+ QW C+G+ + P+A +K IPV
Sbjct: 3037 TWEQWGICIGIAVVSWPLACLVKLIPV 3117
>TC87649 similar to SP|Q9SZR1|ACAA_ARATH Potential calcium-transporting
ATPase 10 plasma membrane-type (EC 3.6.3.8), partial
(43%)
Length = 2159
Score = 358 bits (919), Expect(3) = e-128
Identities = 191/340 (56%), Positives = 230/340 (67%), Gaps = 12/340 (3%)
Frame = +3
Query: 385 KKRMGVVVEQPDGSVRAHCKGASEIILAACDKVIDLNGDVVALDGESTNYLNSIINQFAN 444
K+ GV ++ D V H KGA+EI+LA C ID N +V +D E + I A+
Sbjct: 63 KREAGVAIQTADSDVHIHWKGAAEIVLACCTGYIDANDQLVEIDEEKMTFFKKAIEDMAS 242
Query: 445 EALRTLCLAYMELENGFAAEDP-------IPASGYTCIGIVGIKDPVRPGVKQSVAECRS 497
++LR + +AY E ++ +P + IVGIKDP RPGVK SV C+
Sbjct: 243 DSLRCVAIAYRPYEKEKVPDNEEQLADWSLPEEELVLLAIVGIKDPCRPGVKNSVQLCQK 422
Query: 498 AGIVVRMVTGDNINTAKAIARECGILTD-----DGIAIEGPDFREKTQEELFELIPKIQV 552
AG+ V+MVTGDN+ TAKAIA ECGIL+ + IEG FR + E E+ I V
Sbjct: 423 AGVKVKMVTGDNVKTAKAIALECGILSSLADVTERSVIEGKTFRALSDSEREEIAESISV 602
Query: 553 MARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADV 612
M RSSP DK LV+ LR G VVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKES+D+
Sbjct: 603 MGRSSPNDKLLLVQALRRK-GHVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESSDI 779
Query: 613 IILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNVVALLVNFTSACMTGSAPLTAVQLL 672
IILDDNF+++V V RWGRSVY NIQKF+QFQLTVNV AL++N +A +G PL AVQLL
Sbjct: 780 IILDDNFASVVKVVRWGRSVYANIQKFIQFQLTVNVAALVINVVAAVSSGDVPLNAVQLL 959
Query: 673 WVNMIMDTLGALALATEPPTDDLMKREPVGRKGDFINNVM 712
WVN+IMDTLGALALATEPPTD LM R PVGR+ I N+M
Sbjct: 960 WVNLIMDTLGALALATEPPTDHLMDRSPVGRREPLITNIM 1079
Score = 118 bits (295), Expect(3) = e-128
Identities = 60/131 (45%), Positives = 86/131 (64%), Gaps = 3/131 (2%)
Frame = +1
Query: 713 WRNILGQALYQFVVIWFLQSVGKWVFFLRGP---NADIVLNTLIFNTFVFCQVFNEINSR 769
WRN+L QA+YQ V+ L G + L +A V NTLIFN FV CQ+FNE N+R
Sbjct: 1081 WRNLLIQAMYQVSVLLVLNFRGISILGLEHQPTEHAIKVKNTLIFNAFVICQIFNEFNAR 1260
Query: 770 EMEEIDVFKGIWDNHVFVAVISATVVFQIIIVEYLGTFANTTPLSLVQWIFCLGVGYMGM 829
+ +E ++FKG+ N++F+ ++ TVV Q+IIVE+LG F TT L+ QW+ + +G++G
Sbjct: 1261 KPDEYNIFKGVTRNYLFMGIVGFTVVLQVIIVEFLGKFTTTTRLNWKQWLISVAIGFIGW 1440
Query: 830 PIAVRLKQIPV 840
P+AV K IPV
Sbjct: 1441 PLAVVGKLIPV 1473
Score = 24.3 bits (51), Expect(3) = e-128
Identities = 11/18 (61%), Positives = 12/18 (66%)
Frame = +2
Query: 370 REACKIVKVEPFNSEKKR 387
R I+ V PFNSEKKR
Sbjct: 17 RSESSILHVFPFNSEKKR 70
>TC88464 similar to SP|Q9LU41|ACA9_ARATH Potential calcium-transporting
ATPase 9 plasma membrane-type (EC 3.6.3.8) (Ca2+-ATPase
isoform 9)., partial (43%)
Length = 1608
Score = 451 bits (1159), Expect = e-127
Identities = 243/459 (52%), Positives = 310/459 (66%), Gaps = 14/459 (3%)
Frame = +3
Query: 396 DGSVRAHCKGASEIILAACDKVIDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYM 455
D V H KGA+EI+L AC + +D N + +++ E ++L I+ A +LR + +AY
Sbjct: 6 DSGVHIHWKGAAEIVLGACTQYLDSNSHLQSIEQEK-DFLKEAIDDMAARSLRCVAIAYR 182
Query: 456 ELE-------NGFAAEDPIPASGYTCIGIVGIKDPVRPGVKQSVAECRSAGIVVRMVTGD 508
E A+ +P + IVGIKDP RPGVK +V C AG+ VRMVTGD
Sbjct: 183 SYELDKIPSNEEDLAQWSLPEDELVLLAIVGIKDPCRPGVKDAVRICTDAGVKVRMVTGD 362
Query: 509 NINTAKAIARECGILTD-----DGIAIEGPDFREKTQEELFELIPKIQVMARSSPLDKHT 563
N+ TAKAIA ECGIL D IEG FRE +++E ++ KI VM RSSP DK
Sbjct: 363 NLQTAKAIALECGILASNEEAVDPCIIEGKVFRELSEKEREQVAKKITVMGRSSPNDKLL 542
Query: 564 LVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIV 623
LV+ LR G+VVAVTGDGTNDAPALHEADIGL+MGI GTEVAKES+D+IILDDNF+++V
Sbjct: 543 LVQALRKG-GDVVAVTGDGTNDAPALHEADIGLSMGIQGTEVAKESSDIIILDDNFASVV 719
Query: 624 TVARWGRSVYINIQKFVQFQLTVNVVALLVNFTSACMTGSAPLTAVQLLWVNMIMDTLGA 683
V RWGRSVY NIQKF+QFQLTVNV AL++N +A +G PL AVQLLWVN+IMDTLGA
Sbjct: 720 KVVRWGRSVYANIQKFIQFQLTVNVAALVINVVAAISSGDVPLNAVQLLWVNLIMDTLGA 899
Query: 684 LALATEPPTDDLMKREPVGRKGDFINNVMWRNILGQALYQFVVIWFLQSVGKWVFFLRGP 743
LALATEPPTD LM R PVGR+ I N+MWRN++ QALYQ V+ FL G+ + +
Sbjct: 900 LALATEPPTDHLMHRSPVGRREPLITNIMWRNLIVQALYQVSVLLFLNFCGESILPKQDT 1079
Query: 744 NAD--IVLNTLIFNTFVFCQVFNEINSREMEEIDVFKGIWDNHVFVAVISATVVFQIIIV 801
A V NTLIFN FV CQ+FNE N+R+ +E++VF G+ N +F+ +I T + QI+I+
Sbjct: 1080KAHDYQVKNTLIFNAFVMCQIFNEFNARKPDEMNVFPGVTKNRLFMGIIGITFILQIVII 1259
Query: 802 EYLGTFANTTPLSLVQWIFCLGVGYMGMPIAVRLKQIPV 840
E+LG FA+T L W+ L +G + P+A+ K IPV
Sbjct: 1260EFLGKFASTVRLDWKLWLVSLIIGLVSWPLAIAGKFIPV 1376
>TC92182 type IIA calcium ATPase [Medicago truncatula]
Length = 3148
Score = 339 bits (870), Expect = 2e-93
Identities = 273/853 (32%), Positives = 428/853 (50%), Gaps = 99/853 (11%)
Frame = +1
Query: 1 MTLMILGVCAFVSLIVGVLTEGWPKGAHDGLGIVASILLVVFVTATSDYRQSLQFKDLD- 59
M + IL AF+S ++ EG G + + IL++V ++++ K L+
Sbjct: 190 MLVKILLAAAFISFLLAYF-EGSESGFEAYVEPLVIILILVLNAIVGVWQENNAEKALEA 366
Query: 60 -KEKKKISIQVTRNGYR-QKMSIYNLLPGDIVHLNIGDQVPADGLFVS--GFSVLIDESS 115
KE + SI+V R+GY + L+PGDIV L +GD+VPAD + ++ +++SS
Sbjct: 367 LKELQCESIKVLRDGYFVPDLPARELVPGDIVELRVGDKVPADMRVAALKTSTLRLEQSS 546
Query: 116 LTGESEPIMVTTQNPFL------------LSGTKVQDGSCTMLVTTVGMRTQWGKLMATL 163
LTGE+ P++ T F+ +GT V +GSC +V T M T+ GK+ +
Sbjct: 547 LTGEAMPVLKGTNPIFMDDCELQAKENMVFAGTTVVNGSCICIVITTAMNTEIGKIQKQI 726
Query: 164 SEGG--DDETPLQVKLNG----VATIIGKIGLVFAVITFTVLVKGHLSHKIREGNFWRWT 217
E + +TPL+ KL+ + T IG + LV +I + K +S + +G W
Sbjct: 727 HEASLEESDTPLKKKLDEFGGRLTTSIGIVCLVVWIINY----KNFISWDVVDG----WP 882
Query: 218 GD---NAMEMLEYFAIAVTIVVVAVPEGLPLAVTLSLAFAMKKMMNDKALVRHLAACETM 274
+ + + YF IAV + V A+PEGLP +T LA +KM A+VR L + ET+
Sbjct: 883 TNIQFSFQKCTYYFKIAVALAVAAIPEGLPAVITTCLALGTRKMAQKNAIVRKLPSVETL 1062
Query: 275 GSATTICSDKTGTLTTNRMTVV-------KTCICMNSKEVSNSSSSSDIPDSA------- 320
G T ICSDKTGTLTTN+M+ KT C + +S ++ D D
Sbjct: 1063 GCTTVICSDKTGTLTTNQMSATEFFTLGGKTTAC---RVISVEGTTYDPKDGGIVDWTCY 1233
Query: 321 ---AKLLLQS----IFNNTGGEVVYNKKGKREILGTPTETAILEFGLSLG-GDSKAEREA 372
A LL + + N+ G VY G PTE A+ +G D+K+ +
Sbjct: 1234 NMDANLLAMAEICAVCNDAG---VYFDGRLFRATGLPTEAALKVLVEKMGFPDTKSRNKT 1404
Query: 373 ------------CKIVKVE---------------PFNSEKKRMGVVVEQPDGSVRAHCKG 405
C +K+ F+ +K M V+V +PDG R KG
Sbjct: 1405 HDALVATNNMVDCNTLKLGCCEWWNRRSKRVATLEFDRVRKSMSVIVREPDGQNRLLVKG 1584
Query: 406 ASEIILAACDKVIDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAA-- 463
A E +L V +G +V +D + L +++ +++ LR L LA + F+
Sbjct: 1585 AVESLLERSSYVQLADGSLVPIDDQCRELLLQRLHEMSSKGLRCLGLACKDELGEFSDYY 1764
Query: 464 EDPIPA--------------SGYTCIGIVGIKDPVRPGVKQSVAECRSAGIVVRMVTGDN 509
D PA S +G+VG++DP R V +++ +C+ AGI V ++TGDN
Sbjct: 1765 ADTHPAHKKLLDPTYYSSIESDLIFVGVVGLRDPPREEVHKAIEDCKQAGIRVMVITGDN 1944
Query: 510 INTAKAIARECGILTDD----GIAIEGPDFREKTQEELFELIPKI--QVMARSSPLDKHT 563
+TA+AI +E + + D G ++ G +F + E +L+ + +V +R+ P K
Sbjct: 1945 KSTAEAICKEIKLFSTDEDLTGQSLTGKEFMSLSHSEQVKLLLRNGGKVFSRAEPRHKQE 2124
Query: 564 LVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIV 623
+V+ L+ GE+VA+TGDG NDAPAL ADIG+AMGI GTEVAKE++D+++ DDNFSTIV
Sbjct: 2125 IVRLLKE-MGEIVAMTGDGVNDAPALKLADIGIAMGITGTEVAKEASDMVLADDNFSTIV 2301
Query: 624 TVARWGRSVYINIQKFVQFQLTVNVVALLVNFTSACMTGSAPLTAVQLLWVNMIMDTLGA 683
+ GR++Y N++ F+++ ++ NV ++ F +A + + VQLLWVN++ D A
Sbjct: 2302 SAIAEGRAIYNNMKAFIRYMISSNVGEVISIFLTAALGIPECMIPVQLLWVNLVTDGPPA 2481
Query: 684 LALATEPPTDDLMKREPVGRKGDFINNVMWRNILGQALYQFVVI-WFL-QSVGKWVFFLR 741
AL P D+M++ P RK D W L++++V WFL ++ W F
Sbjct: 2482 TALGFNPADVDIMQKPP--RKSDDALISAW------VLFRYLVSNWFLCRNCNSWHFC-- 2631
Query: 742 GPNADIVLNTLIF 754
IV++T IF
Sbjct: 2632 -----IVVHTSIF 2655
>CB894102 similar to GP|8843813|db Ca2+-transporting ATPase-like protein
{Arabidopsis thaliana}, partial (15%)
Length = 802
Score = 191 bits (486), Expect = 8e-49
Identities = 93/165 (56%), Positives = 132/165 (79%)
Frame = +2
Query: 36 SILLVVFVTATSDYRQSLQFKDLDKEKKKISIQVTRNGYRQKMSIYNLLPGDIVHLNIGD 95
+++LV+ VTA SDY+QSLQF+DL++EK+ I ++V R G R ++SIY+L+ GD++ LNIG+
Sbjct: 5 AVILVIVVTAVSDYKQSLQFRDLNEEKRNIHLEVIRGGRRVEISIYDLVVGDVIPLNIGN 184
Query: 96 QVPADGLFVSGFSVLIDESSLTGESEPIMVTTQNPFLLSGTKVQDGSCTMLVTTVGMRTQ 155
QVPADG+ ++G S+ IDESS+TGES+ + +++PF++SG KV DGS TMLVT VG+ T+
Sbjct: 185 QVPADGVVITGHSLSIDESSMTGESKIVHKDSKDPFMMSGCKVADGSGTMLVTGVGINTE 364
Query: 156 WGKLMATLSEGGDDETPLQVKLNGVATIIGKIGLVFAVITFTVLV 200
WG LMA++SE +ETPLQV+LNGVAT IG +GL AV+ VL+
Sbjct: 365 WGLLMASISEDTGEETPLQVRLNGVATFIGIVGLSVAVLVLIVLL 499
>BF004483 GP|21314227| type IIB calcium ATPase MCA5 {Medicago truncatula},
partial (11%)
Length = 551
Score = 181 bits (460), Expect = 8e-46
Identities = 87/109 (79%), Positives = 96/109 (87%)
Frame = +1
Query: 654 NFTSACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKREPVGRKGDFINNVMW 713
N +G+APLTAVQLLWVNMIMDTLGALALATEPP DDLMKR PVGRKG+FI+NVMW
Sbjct: 46 NIICILSSGTAPLTAVQLLWVNMIMDTLGALALATEPPNDDLMKRAPVGRKGNFISNVMW 225
Query: 714 RNILGQALYQFVVIWFLQSVGKWVFFLRGPNADIVLNTLIFNTFVFCQV 762
RNILGQ+LYQF+VIWFLQS GK +F L GPN+D+VLNTLIFN FVFCQV
Sbjct: 226 RNILGQSLYQFMVIWFLQSKGKTIFSLDGPNSDLVLNTLIFNAFVFCQV 372
Score = 30.0 bits (66), Expect = 3.9
Identities = 13/15 (86%), Positives = 15/15 (99%)
Frame = +2
Query: 761 QVFNEINSREMEEID 775
QVFNEINSREME+I+
Sbjct: 506 QVFNEINSREMEKIN 550
>TC78163 H+-ATPase
Length = 3295
Score = 173 bits (439), Expect = 2e-43
Identities = 150/589 (25%), Positives = 261/589 (43%), Gaps = 4/589 (0%)
Frame = +1
Query: 68 QVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGESEPIMVTT 127
+V R+G + L+PGDIV + +GD +PAD + G + ID+S+LTGES P+ T
Sbjct: 553 KVLRDGKWSEEDASVLVPGDIVSIKLGDIIPADARLLEGDPLKIDQSALTGESLPV---T 723
Query: 128 QNPF--LLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATIIG 185
++P + SG+ + G +V G+ T +GK A L E Q L +
Sbjct: 724 KHPGEGIYSGSTCKQGEIEAIVIATGVHTFFGKA-AHLVENTTHVGHFQKVLTSIGNFCI 900
Query: 186 KIGLVFAVITFTVLVKGHLSHKIREGNFWRWTGDNAMEMLEYFAIAVTIVVVAVPEGLPL 245
+ VI V+ H G +R DN + +L + +P +P
Sbjct: 901 CSIAIGMVIEIIVIYGVH-------GKGYRNGIDNLLVLL----------IGGIPIAMPT 1029
Query: 246 AVTLSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNRMTVVKTCICMNSK 305
+++++A K+ A+ + + A E M +CSDKTGTLT N++TV K I + +K
Sbjct: 1030 VLSVTMAIGSHKLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLTVDKDMIEVFAK 1209
Query: 306 EVSNSSSSSDIPDSAAKLLLQSIFNNTGGEVVYNKKGKREILGTPTETAILEFGLSLGGD 365
V V+ + R + AI+ S+ D
Sbjct: 1210 GVDKDLV-----------------------VLMAARASRLENQDAIDCAIV----SMLAD 1308
Query: 366 SKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSVRAHCKGASEIILAACDKVIDLNGDVV 425
K R K V PFN KR + G++ KGA E IL ++
Sbjct: 1309 PKEARTGIKEVHFLPFNPTDKRTALTYIDAAGNMHRVSKGAPEQIL-----------NLA 1455
Query: 426 ALDGESTNYLNSIINQFANEALRTLCLAYMELENGFAAEDPIPASGYTCIGIVGIKDPVR 485
E ++S+I++FA LR+L +A E+ G P + + ++ + DP R
Sbjct: 1456 RNKAEIAQKVHSMIDKFAERGLRSLGVARQEVPEG---SKDSPGGPWEFVALLPLFDPPR 1626
Query: 486 PGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTD--DGIAIEGPDFREKTQEEL 543
+++ G+ V+M+TGD + K R G+ T+ ++ G + + +
Sbjct: 1627 HDSAETIRRALDLGVSVKMITGDQLAIGKETGRRLGMGTNMYPSSSLLGDNKDQLGAVSI 1806
Query: 544 FELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGT 603
+LI A P K+ +VK+L+ + +TGDG NDAPAL ADIG+A+ T
Sbjct: 1807 DDLIENADGFAGVFPEHKYEIVKRLQAR-KHICGMTGDGVNDAPALKIADIGIAVA-DST 1980
Query: 604 EVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNVVALL 652
+ A+ ++D+++ + S I++ R+++ ++ + + +++ + +L
Sbjct: 1981 DAARSASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVL 2127
>BG647796 GP|21314227|g type IIB calcium ATPase MCA5 {Medicago truncatula},
partial (9%)
Length = 780
Score = 166 bits (420), Expect = 3e-41
Identities = 80/97 (82%), Positives = 90/97 (92%)
Frame = +1
Query: 744 NADIVLNTLIFNTFVFCQVFNEINSREMEEIDVFKGIWDNHVFVAVISATVVFQIIIVEY 803
++D+VLNTLIFN FVFCQVFNEINSREME+I+VFKGI DN+VFV VISAT+ FQIIIVEY
Sbjct: 1 DSDLVLNTLIFNAFVFCQVFNEINSREMEKINVFKGILDNYVFVGVISATIFFQIIIVEY 180
Query: 804 LGTFANTTPLSLVQWIFCLGVGYMGMPIAVRLKQIPV 840
LGTFANTTPL+LVQW FCL VG+MGMPIA RLK+IPV
Sbjct: 181 LGTFANTTPLTLVQWFFCLFVGFMGMPIAARLKKIPV 291
>TC77796 H+-ATPase
Length = 3416
Score = 117 bits (292), Expect(2) = 2e-40
Identities = 96/373 (25%), Positives = 168/373 (44%), Gaps = 4/373 (1%)
Frame = +1
Query: 280 ICSDKTGTLTTNRMTVVKTCICMNSKEVSNSSSSSDIPDSAAKLLLQSIFNNTGGEVVYN 339
+CSDKTGTLT N+++V K I + +K V ++
Sbjct: 1075 LCSDKTGTLTLNKLSVEKNLIEVFAKGVEKDYV-----------------------ILLA 1185
Query: 340 KKGKREILGTPTETAILEFGLSLGGDSKAEREACKIVKVEPFNSEKKRMGVVVEQPDGSV 399
+ R T + AI + + D K R + V PFN KR + DG+
Sbjct: 1186 ARASR----TENQDAIDAAIVGMLADPKEARAGVREVHFFPFNPVDKRTALTYIDADGNW 1353
Query: 400 RAHCKGASEIILAACDKVIDLNGDVVALDGESTNYLNSIINQFANEALRTLCLAYMELEN 459
KGA E IL C+ D+ +S I++FA LR+L +A E+
Sbjct: 1354 HRSSKGAPEQILNLCNCKEDVRKKA-----------HSTIDKFAERGLRSLGVARQEIPE 1500
Query: 460 GFAAEDPIPASGYTCIGIVGIKDPVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARE 519
+ P + + +G++ + DP R +++ + G+ V+M+TGD + AK R
Sbjct: 1501 ---KDKDSPGAPWQFVGLLPLFDPPRHDSAETITRALNLGVNVKMITGDQLAIAKETGRR 1671
Query: 520 CGILTD----DGIAIEGPDFREKTQEELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEV 575
G+ T+ + + D + ELI K A P K+ +VK+L+ +
Sbjct: 1672 LGMGTNMYPSSSLLGQSKDAAVSALP-VDELIEKADGFAGVFPEHKYEIVKKLQER-KHI 1845
Query: 576 VAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYIN 635
+TGDG NDAPAL ADIG+A+ A T+ A+ ++D+++ + S I++ R+++
Sbjct: 1846 CGMTGDGVNDAPALKRADIGIAVADA-TDAARGASDIVLTEPGLSVIISAVLTSRAIFQR 2022
Query: 636 IQKFVQFQLTVNV 648
++ + + +++ +
Sbjct: 2023 MKNYTIYAVSITI 2061
Score = 68.2 bits (165), Expect(2) = 2e-40
Identities = 57/212 (26%), Positives = 99/212 (45%), Gaps = 5/212 (2%)
Frame = +3
Query: 68 QVTRNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGESEPIMVTT 127
+V R+G + L+PGDI+ + +GD +PAD + G ++ +D+S+LTGES P T
Sbjct: 495 RVLRDGRWSEEDAAILVPGDIISIKLGDIIPADARLLEGDALSVDQSALTGESLP---AT 665
Query: 128 QNPF--LLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATIIG 185
+NP + SG+ V+ G +V G+ T +GK + D T V T IG
Sbjct: 666 KNPSDEVFSGSTVKKGEIEAVVIATGVHTFFGKAAHLV-----DSTNQVGHFQKVLTAIG 830
Query: 186 K--IGLVFAVITFTVLVKGHLSH-KIREGNFWRWTGDNAMEMLEYFAIAVTIVVVAVPEG 242
I + I ++V + H K R+G DN + +L + +P
Sbjct: 831 NFCICSIAVGILIELVVMYPIQHRKYRDGI------DNLLVLL----------IGGIPIA 962
Query: 243 LPLAVTLSLAFAMKKMMNDKALVRHLAACETM 274
+P +++++A ++ A+ + + A E M
Sbjct: 963 MPTVLSVTMAIGSHRLSQQGAITKRMTAIEEM 1058
>TC79651 similar to PIR|T52581|T52581 Ca2+-transporting ATPase (EC 3.6.1.38)
ECA3 [imported] - Arabidopsis thaliana, partial (38%)
Length = 1546
Score = 136 bits (343), Expect = 3e-32
Identities = 107/359 (29%), Positives = 176/359 (48%), Gaps = 32/359 (8%)
Frame = +3
Query: 507 GDNINTAKAIARECGILTDDGIAIEGPDFREKTQEELFEL-----IPKIQVMARSSPLDK 561
G+N +TA+++ R+ G D I + EEL L + ++ + R P K
Sbjct: 18 GNNKSTAESLCRKIGAF-DHLIDFTEHSYTASEFEELPALQQTIALQRMALFTRVEPSHK 194
Query: 562 HTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFST 621
LV+ L+ EVVA+TGDG NDAPAL +ADIG+AMG +GT VAK ++D+++ DDNF++
Sbjct: 195 RMLVEALQHQ-NEVVAMTGDGVNDAPALKKADIGIAMG-SGTAVAKSASDMVLADDNFAS 368
Query: 622 IVTVARWGRSVYINIQKFVQFQLTVNVVALLVNFTSACMTGSAPLTAVQLLWVNMIMDTL 681
IV GR++Y N ++F+++ ++ N+ ++ F +A + L VQLLWVN++ D L
Sbjct: 369 IVAAVAEGRAIYNNTKQFIRYMISSNIGEVVCIFVAAVLGIPDTLAPVQLLWVNLVTDGL 548
Query: 682 GALALATEPPTDDLMKREPVGRKGD--------FINNVMWRNILGQALYQFVVIWFLQS- 732
A A+ D+MK +P RK + F ++ +G A + WF+ +
Sbjct: 549 PATAIGFNKQDSDVMKVKP--RKVNEAVVTGWLFFRYLVIGAYVGLATVAGFIWWFVYAD 722
Query: 733 VGKWVFFLRGPNADIV----------------LNTLIFNTFVFCQVFNEINSREMEEIDV 776
G + + N D +T+ V ++FN +N+ + +
Sbjct: 723 SGPKLPYTELMNFDTCPTRETTYPCSIFEDRHPSTVAMTVLVVVEMFNALNNLSENQSLL 902
Query: 777 FKGIWDNHVFVAVISATVVFQIII--VEYLGTFANTTPLSLVQWIFCLGVGYMGMPIAV 833
W N V I T++ I+I V L + TPLS W + V Y+ +P+ +
Sbjct: 903 VIPPWSNLWLVGSIVLTMLLHILILYVRPLSVLFSVTPLSWADW---MAVLYLSLPVII 1070
>TC86961 similar to GP|1742951|emb|CAA70946.1 Ca2+-ATPase {Arabidopsis
thaliana}, partial (77%)
Length = 1488
Score = 130 bits (328), Expect = 2e-30
Identities = 95/299 (31%), Positives = 143/299 (47%), Gaps = 52/299 (17%)
Frame = +1
Query: 578 VTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQ 637
+TGDG NDAPAL ADIG+AMGIAGTEVAKE++D+++ DDNFS+IV GRS+Y N++
Sbjct: 55 MTGDGVNDAPALKLADIGIAMGIAGTEVAKEASDMVLADDNFSSIVAAVGEGRSIYNNMK 234
Query: 638 KFVQFQLTVNVVALLVNFTSACMTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMK 697
F+++ ++ N+ + F +A + L VQLLWVN++ D A AL PP D+MK
Sbjct: 235 AFIRYMISSNIGEVASIFLTAALGIPEGLIPVQLLWVNLVTDGPPATALGFNPPDKDIMK 414
Query: 698 REPVGRKGDFINN-VMWRNI-----LGQALYQFVVIWF---------LQSVG-------- 734
+ P IN +++R + +G A +IW+ L S G
Sbjct: 415 KPPRRSDDSLINLWILFRYLVIGIYVGLATVGVFIIWYTHGSFMGIDLSSDGHTLVTYSQ 594
Query: 735 --------KWVFFLRGP----------NAD---------IVLNTLIFNTFVFCQVFNEIN 767
W F P +AD + TL + V ++FN +N
Sbjct: 595 LANWGQCSSWNNFTAAPFTAGSRIISFDADPCDYFTTGKVKAMTLSLSVLVAIEMFNSLN 774
Query: 768 SREMEEIDVFKGIWDNHVFVAVISAT--VVFQIIIVEYLGTFANTTPLSLVQWIFCLGV 824
+ + + W N + +S + + F I+ V +L PLS +W+ L V
Sbjct: 775 ALSEDGSLLTMPPWVNPWLLLAMSVSFGLHFLILYVPFLAKVFGIVPLSFNEWLLVLAV 951
>CB893893 similar to GP|8843813|db Ca2+-transporting ATPase-like protein
{Arabidopsis thaliana}, partial (11%)
Length = 645
Score = 126 bits (317), Expect = 3e-29
Identities = 72/150 (48%), Positives = 102/150 (68%), Gaps = 3/150 (2%)
Frame = +1
Query: 71 RNGYRQKMSIYNLLPGDIVHLNIGDQVPADGLFVSGFSVLIDESSLTGESEPIMVTTQNP 130
R G R ++SIY+L+ GD++ LNIG+QVPADG+ ++G S+ IDESS+TGES+ + +++P
Sbjct: 4 RGGRRVEISIYDLVVGDVIPLNIGNQVPADGVVITGHSLSIDESSMTGESKIVHKDSKDP 183
Query: 131 FLLSGTKVQDGSCTMLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATIIGKIGLV 190
F++SG KV DGS TMLVT VG+ T+WG LMA++SE +ETPLQV + L+
Sbjct: 184 FMMSGCKVADGSGTMLVTGVGINTEWGLLMASISEDTGEETPLQVFF---LAAVSDCVLL 354
Query: 191 FAVITFT--VLVKGHLSHKIREGNFW-RWT 217
F +I VL+ G L + R +W RWT
Sbjct: 355 FLIIQLPL*VLIIGPL--EWRSHLYWYRWT 438
Score = 31.6 bits (70), Expect = 1.3
Identities = 15/26 (57%), Positives = 19/26 (72%)
Frame = +2
Query: 175 VKLNGVATIIGKIGLVFAVITFTVLV 200
V+LNGVAT IG +GL AV+ VL+
Sbjct: 395 VRLNGVATFIGIVGLSVAVLVLIVLL 472
>CB892538 similar to SP|Q9XES1|ECA Calcium-transporting ATPase 4 endoplasmic
reticulum-type (EC 3.6.3.8). [Mouse-ear cress], partial
(24%)
Length = 805
Score = 90.9 bits (224), Expect(2) = 1e-28
Identities = 63/185 (34%), Positives = 98/185 (52%), Gaps = 22/185 (11%)
Frame = +2
Query: 381 FNSEKKRMGVVVEQPDGSVRAHC-KGASEIILAACDKVIDLNGDVVALDGESTNYLNSII 439
F+ ++K MGV+V+ G ++ KGA E +L KV +G VV LD + N + +
Sbjct: 59 FDRDRKSMGVIVDSGVGKKKSLLVKGAVENVLDRSSKVQLRDGSVVKLDNNAKNLILQAL 238
Query: 440 NQFANEALRTLCLAYME----LENGFAAEDP------IPASGYTCI-------GIVGIKD 482
++ + ALR L AY + EN ED + + Y+ I G+VG++D
Sbjct: 239 HEMSTSALRCLGFAYKDELTNFENYNGNEDHPAHQLLLDPNNYSSIEDELIFVGLVGLRD 418
Query: 483 PVRPGVKQSVAECRSAGIVVRMVTGDNINTAKAIARECGILTDD----GIAIEGPDFREK 538
P R V Q++ +CR+AGI V ++TGDN NTA+AI RE G+ + ++ G DF E
Sbjct: 419 PPREEVYQAIEDCRAAGIRVMVITGDNKNTAEAICREIGVFAPNENISSKSLTGKDFMEL 598
Query: 539 TQEEL 543
++L
Sbjct: 599 RDKKL 613
Score = 55.1 bits (131), Expect(2) = 1e-28
Identities = 28/35 (80%), Positives = 30/35 (85%)
Frame = +3
Query: 573 GEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAK 607
GEVVA+TGDG NDA AL ADIG+AMGIA TEVAK
Sbjct: 699 GEVVAMTGDGCNDATALKLADIGIAMGIARTEVAK 803
>BQ138304 homologue to GP|6688833|emb putative calcium P-type ATPase
{Neurospora crassa}, partial (13%)
Length = 412
Score = 113 bits (282), Expect = 3e-25
Identities = 61/138 (44%), Positives = 90/138 (65%)
Frame = +3
Query: 540 QEELFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMG 599
+ E E + +R+ P K LV L+ GEVVA+TGDG NDAPAL +ADIG+AMG
Sbjct: 3 EAEQMEAAKNASLFSRTEPTHKSKLVDLLQKA-GEVVAMTGDGVNDAPALKKADIGVAMG 179
Query: 600 IAGTEVAKESADVIILDDNFSTIVTVARWGRSVYINIQKFVQFQLTVNVVALLVNFTSAC 659
+GT+VAK +AD++++DDNF+TI GRS+Y N Q+F+++ ++ N+ ++ F +A
Sbjct: 180 -SGTDVAKLAADMVLVDDNFATIEGAVEEGRSIYNNTQQFIRYLISSNIGEVVSIFLTAA 356
Query: 660 MTGSAPLTAVQLLWVNMI 677
L VQLLWVN++
Sbjct: 357 AGMPEALIPVQLLWVNLV 410
>AI974405 similar to SP|Q9LF79|ACA Calcium-transporting ATPase 8 plasma
membrane-type (EC 3.6.3.8) (Ca2+-ATPase isoform 8).,
partial (10%)
Length = 332
Score = 93.6 bits (231), Expect = 3e-19
Identities = 55/110 (50%), Positives = 71/110 (64%), Gaps = 7/110 (6%)
Frame = +2
Query: 163 LSEGGDDETPLQVKLNGVATIIGKIGLVFAVITFTVLVKGHLSHKIREGNFWRW------ 216
+SE +ETPLQV+LNGVAT IG +GL AV+ VL+ + S R + +
Sbjct: 2 ISEDTGEETPLQVRLNGVATFIGIVGLSVAVLVLIVLLARYFSGHTRNSDGTKQFIAGKT 181
Query: 217 -TGDNAMEMLEYFAIAVTIVVVAVPEGLPLAVTLSLAFAMKKMMNDKALV 265
G ++ +AVTIVVVAVPEGLPLAVTL+LA++M+KMM DKALV
Sbjct: 182 KAGHAIDGAIKIITVAVTIVVVAVPEGLPLAVTLTLAYSMRKMMADKALV 331
>CB893927 homologue to GP|17342714|g type IIA calcium ATPase {Medicago
truncatula}, partial (13%)
Length = 413
Score = 82.8 bits (203), Expect(2) = 1e-18
Identities = 48/116 (41%), Positives = 74/116 (63%), Gaps = 6/116 (5%)
Frame = +2
Query: 496 RSAGIVVRMVTGDNINTAKAIARECGILTDD----GIAIEGPDFREKTQEELFELIPKI- 550
+ AGI V ++TG+N +TA+AI +E + + D G ++ G +F + E +L+ +
Sbjct: 2 KQAGIRVMVITGENKSTAEAICKEIKLFSTDEDLTGQSLTGKEFMSLSHSEQVKLLLRNG 181
Query: 551 -QVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEV 605
+ +R+ P K +V+ L+ GE+VA+TGDG NDAPAL ADIG+AMGI GTE+
Sbjct: 182 GKGFSRAEPRHKQEIVRLLKE-MGEIVAMTGDGVNDAPALKLADIGMAMGITGTEM 346
Score = 29.3 bits (64), Expect(2) = 1e-18
Identities = 12/22 (54%), Positives = 17/22 (76%)
Frame = +1
Query: 616 DDNFSTIVTVARWGRSVYINIQ 637
DDNFSTIV+ GR++Y N++
Sbjct: 343 DDNFSTIVSAIAEGRAIYNNMK 408
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.137 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,466,350
Number of Sequences: 36976
Number of extensions: 281230
Number of successful extensions: 1464
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 1404
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1423
length of query: 840
length of database: 9,014,727
effective HSP length: 104
effective length of query: 736
effective length of database: 5,169,223
effective search space: 3804548128
effective search space used: 3804548128
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC147000.2


