

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146972.3 + phase: 0
(240 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
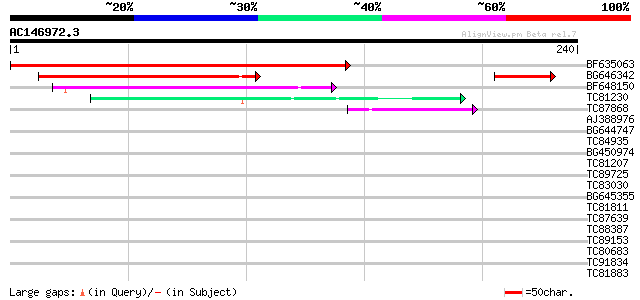
BF635063 weakly similar to PIR|F84486|F84 probable retroelement ... 261 1e-70
BG646342 weakly similar to PIR|F84486|F84 probable retroelement ... 139 1e-33
BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x pa... 65 2e-11
TC81230 41 4e-04
TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 40 8e-04
AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing fact... 39 0.002
BG644747 37 0.009
TC84935 similar to PIR|G96631|G96631 probable RNA-binding protei... 36 0.015
BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR p... 36 0.015
TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 35 0.020
TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.... 34 0.044
TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA... 32 0.22
BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.... 31 0.49
TC81811 similar to GP|6671365|gb|AAF23176.1| P-glycoprotein {Gos... 31 0.49
TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.... 30 0.83
TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finge... 28 2.4
TC89153 similar to GP|18855061|gb|AAL79753.1 putative RNA helica... 28 2.4
TC80683 homologue to GP|8777424|dbj|BAA97014.1 gb|AAF56406.1~gen... 28 3.2
TC91834 similar to PIR|T08416|T08416 disease resistance protein ... 28 3.2
TC81883 weakly similar to GP|7110148|gb|AAF36810.1| DNA repair-r... 28 4.1
>BF635063 weakly similar to PIR|F84486|F84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(4%)
Length = 677
Score = 261 bits (668), Expect = 1e-70
Identities = 136/144 (94%), Positives = 140/144 (96%)
Frame = -2
Query: 1 MSQAEKTEMVDKARSAIVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQ 60
MS+AEKTEMVDKARSAIVLCLGDKVLREVAKE TAASMWAKL SLYMTKSLAHRQFLKQQ
Sbjct: 433 MSRAEKTEMVDKARSAIVLCLGDKVLREVAKERTAASMWAKL*SLYMTKSLAHRQFLKQQ 254
Query: 61 LYSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKNTMLYG 120
LYSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDE+KAILLLCALPKSFESFK+TMLYG
Sbjct: 253 LYSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDEEKAILLLCALPKSFESFKDTMLYG 74
Query: 121 KEGTVTLEEIQAALRTKELTNSKD 144
KEGTVTLEE+QAALRTKELT S D
Sbjct: 73 KEGTVTLEEVQAALRTKELTKSND 2
>BG646342 weakly similar to PIR|F84486|F84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(4%)
Length = 599
Score = 139 bits (349), Expect = 1e-33
Identities = 75/94 (79%), Positives = 80/94 (84%)
Frame = +2
Query: 13 ARSAIVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAI 72
ARSAIVLCLGDKVLREVAKE TA SM AKLE LYMTKSLAHRQFLKQQLYSF+MVESKAI
Sbjct: 2 ARSAIVLCLGDKVLREVAKEPTATSMCAKLEYLYMTKSLAHRQFLKQQLYSFKMVESKAI 181
Query: 73 MEQLTEFNKILDDLENIEVQLEDEDKAILLLCAL 106
E L EFNKI+ DLENIEV LED A+++ C L
Sbjct: 182 TELLVEFNKIIGDLENIEVHLEDAG-ALMVWCCL 280
Score = 57.4 bits (137), Expect = 5e-09
Identities = 24/26 (92%), Positives = 25/26 (95%)
Frame = +2
Query: 206 EDVGALMVWCCLEDEEGDVSHLGSDA 231
ED GALMVWCCLEDEEGDVSHLG+DA
Sbjct: 245 EDAGALMVWCCLEDEEGDVSHLGNDA 322
>BF648150 similar to GP|14586969|gb| pol polyprotein {Citrus x paradisi},
partial (3%)
Length = 658
Score = 65.5 bits (158), Expect = 2e-11
Identities = 37/127 (29%), Positives = 69/127 (54%), Gaps = 7/127 (5%)
Frame = +2
Query: 19 LCLG-------DKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKA 71
+CLG D + +A +W KLE+ YM + ++FL +++MV++K+
Sbjct: 197 ICLGHILNGMSDSLFDIYQSSPSAKDLWDKLETRYMREDATSKKFLVSHFNNYKMVDNKS 376
Query: 72 IMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKNTMLYGKEGTVTLEEIQ 131
+MEQL E +IL++ + + +++ ++ LP S++ FK TM + KE ++LE++
Sbjct: 377 VMEQLYEIERILNNYKQHNMNMDETIIVSSIIDKLPPSWKDFKRTMKHKKE-DISLEQLG 553
Query: 132 AALRTKE 138
LR E
Sbjct: 554 NHLRLXE 574
>TC81230
Length = 958
Score = 41.2 bits (95), Expect = 4e-04
Identities = 39/171 (22%), Positives = 68/171 (38%), Gaps = 12/171 (7%)
Frame = +1
Query: 35 AASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLTEFNKILDDLENIEVQLE 94
A +W L+ Y L+H+ L + L + + + + E L + I + L + E L+
Sbjct: 475 AKEVWDHLKQRYTISDLSHQYQLLKDLSNLKQQSGQPVYEFLAQMEVIWNQLTSCEPSLK 654
Query: 95 DED------------KAILLLCALPKSFESFKNTMLYGKEGTVTLEEIQAALRTKELTNS 142
D + I L AL +E + + L+ + TLE L+++E T
Sbjct: 655 DATDMKTYETHRNRVRLIQFLMALTDEYEPVRASSLH-QNPLPTLENALPCLKSEE-TRL 828
Query: 143 KDLTHEHDEGLSVSRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPEI 193
+ + + D +V+ N + C +C K GH DCP I
Sbjct: 829 QLVPPKADLAFAVT--------------NNATKPCRHCQKSGHSFSDCPTI 939
>TC87868 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (91%)
Length = 860
Score = 40.0 bits (92), Expect = 8e-04
Identities = 19/55 (34%), Positives = 26/55 (46%)
Frame = +3
Query: 144 DLTHEHDEGLSVSRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPEINGNSA 198
+L+H G G GGGRG G S +C+ C + GHF ++C G A
Sbjct: 240 ELSHNSRSG-GGGGGGGGGRGRGGGGGGGSDLKCYECGEPGHFARECRNRGGGGA 401
>AJ388976 similar to PIR|E84638|E84 probable RSZp22 splicing factor
[imported] - Arabidopsis thaliana, partial (62%)
Length = 508
Score = 38.5 bits (88), Expect = 0.002
Identities = 16/40 (40%), Positives = 20/40 (50%)
Frame = +2
Query: 158 GNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPEINGNS 197
G GGG G R G S +C+ C + GHF + C G S
Sbjct: 287 GGGGGGGRGRSGGGGSDLKCYXCGEPGHFARXCNSSPGGS 406
>BG644747
Length = 685
Score = 36.6 bits (83), Expect = 0.009
Identities = 25/104 (24%), Positives = 52/104 (49%), Gaps = 1/104 (0%)
Frame = +1
Query: 12 KARSAIVLCLGDKVLREVAKE-ATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESK 70
K R I CL D + +++ +W L+S+Y + ++ + F+MV++K
Sbjct: 247 KCRYHIFKCLYDNFYDYYDRTYSSSKKIWKALQSMYDIEDARA*KYTDS*FFRFKMVDNK 426
Query: 71 AIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFK 114
++++Q +F I+ L + EV++ D ++ LP S + F+
Sbjct: 427 SMVDQAQDFIMIVRYLRSKEVKIGDNLIVCGIVDKLPPS*KKFQ 558
>TC84935 similar to PIR|G96631|G96631 probable RNA-binding protein F8A5.17
[imported] - Arabidopsis thaliana, partial (41%)
Length = 552
Score = 35.8 bits (81), Expect = 0.015
Identities = 13/41 (31%), Positives = 22/41 (52%)
Frame = +2
Query: 159 NGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPEINGNSAQ 199
+ GGRG+ + +CF C + GH+ +DCP G+ +
Sbjct: 419 SSGGRGSYGAGDRVGQDDCFKCGRPGHWARDCPLAGGDGGR 541
>BG450974 similar to PIR|T05112|T05 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana, partial (54%)
Length = 364
Score = 35.8 bits (81), Expect = 0.015
Identities = 13/33 (39%), Positives = 18/33 (54%)
Frame = +1
Query: 158 GNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDC 190
G GGGRG R +C+ C + GHF ++C
Sbjct: 262 GGGGGRGGGRGGRGGDDLKCYECGEPGHFAREC 360
>TC81207 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (39%)
Length = 630
Score = 35.4 bits (80), Expect = 0.020
Identities = 20/61 (32%), Positives = 26/61 (41%), Gaps = 4/61 (6%)
Frame = +3
Query: 140 TNSKDLTHEHDEGLSVSR----GNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPEING 195
T + D+T E L V + G GGGRG R C+ C GH +DC +
Sbjct: 228 TKAVDVTGPKGEPLQVRQDNHGGGGGGRGFRGGERRNGGGGCYTCGDTGHIARDCDRSDR 407
Query: 196 N 196
N
Sbjct: 408 N 410
Score = 34.7 bits (78), Expect = 0.034
Identities = 25/80 (31%), Positives = 33/80 (41%)
Frame = +3
Query: 157 RGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPEINGNSAQIVYEGYEDVGALMVWCC 216
R GGG G+R ++ C+ C HF +DC GN+ GY G C
Sbjct: 423 RSGGGGGGDRDRA-------CYTCGSFEHFARDCMRGGGNNNN-GGGGYGGGGTSCYRC- 575
Query: 217 LEDEEGDVSHLGSDACNTPN 236
G V H+ D C TP+
Sbjct: 576 -----GGVGHIARD-CATPS 617
>TC89725 similar to PIR|T05494|T05494 glycine-rich protein T19K4.150 -
Arabidopsis thaliana, partial (17%)
Length = 378
Score = 34.3 bits (77), Expect = 0.044
Identities = 15/37 (40%), Positives = 19/37 (50%), Gaps = 3/37 (8%)
Frame = +1
Query: 158 GNGGGR---GNRRKSGNKSRFECFNCHKMGHFKKDCP 191
G GGGR G G C++C + GHF +DCP
Sbjct: 70 GGGGGRYGGGGGGGGGGGGGGSCYSCGESGHFARDCP 180
>TC83030 weakly similar to GP|18855061|gb|AAL79753.1 putative RNA helicase
{Oryza sativa}, partial (7%)
Length = 624
Score = 32.0 bits (71), Expect = 0.22
Identities = 15/39 (38%), Positives = 20/39 (50%), Gaps = 3/39 (7%)
Frame = +1
Query: 161 GGRGNRRKSGNKSRF---ECFNCHKMGHFKKDCPEINGN 196
GGR + R S + +R CF C + GH DCP G+
Sbjct: 109 GGRQSSRSSSSPNRSFAGTCFTCGESGHRASDCPNKRGD 225
>BG645355 similar to PIR|G96590|G965 hypothetical protein T24C10.5 [imported]
- Arabidopsis thaliana, partial (5%)
Length = 627
Score = 30.8 bits (68), Expect = 0.49
Identities = 13/40 (32%), Positives = 21/40 (52%)
Frame = -1
Query: 155 VSRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPEIN 194
VS G+GG GN C+ C++ GH+ +CP ++
Sbjct: 435 VSGGSGGASGN-----------CYKCNQPGHWANNCPNMS 349
Score = 26.9 bits (58), Expect = 7.0
Identities = 9/35 (25%), Positives = 17/35 (47%)
Frame = -1
Query: 160 GGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPEIN 194
GG N + +C+ C + GH+ +CP ++
Sbjct: 552 GGAYVNTVSGSGGASGKCYKCQQPGHWASNCPSMS 448
>TC81811 similar to GP|6671365|gb|AAF23176.1| P-glycoprotein {Gossypium
hirsutum}, partial (17%)
Length = 850
Score = 30.8 bits (68), Expect = 0.49
Identities = 28/94 (29%), Positives = 41/94 (42%), Gaps = 1/94 (1%)
Frame = +1
Query: 82 ILDDLENIEVQLEDEDKAILLLCALPKSFE-SFKNTMLYGKEGTVTLEEIQAALRTKELT 140
++D + + L+ K I L+ P F S +LYGKEG E I+AA +L
Sbjct: 106 LIDGKDITRINLKSLTKHIGLVQQEPALFATSIYENILYGKEGASDSEVIEAA----KLA 273
Query: 141 NSKDLTHEHDEGLSVSRGNGGGRGNRRKSGNKSR 174
N+ + EG S G RG + G + R
Sbjct: 274 NAHNFISALPEGYSTKVGE---RGVQLSGGQRQR 366
>TC87639 GP|9663153|emb|CAC01132.1 transport-secretion protein 2.2 (TTS-2.2)
{Homo sapiens}, partial (2%)
Length = 1522
Score = 30.0 bits (66), Expect = 0.83
Identities = 33/176 (18%), Positives = 68/176 (37%), Gaps = 25/176 (14%)
Frame = -1
Query: 40 AKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLTEFNKILDD-----------LEN 88
A L+ Y+ ++ R QL S R +++ + L F K+L D +
Sbjct: 778 AYLDRTYLDPNIQSRAVA--QLQSLRQKDTERLATFLPRFEKVLADAGGYSWPDVVQISL 605
Query: 89 IEVQLEDEDKAILLLCALPKSFESFKNTMLYGKEGTVTLEEIQAALRTKELTNSKDLTHE 148
+E L K +L+ LP + + + + ++ +E ++ ++ +
Sbjct: 604 LETALVPRLKELLITVELPTVYSQWLSKV---QDIAWKMERMKTPPTRWAPATRLPVSKD 434
Query: 149 HDEGLSVSRGNGGGRGNRRKSGNKSRF--------------ECFNCHKMGHFKKDC 190
D + ++ G RR+ G+ S EC++CH+ GH ++C
Sbjct: 433 RDGDMMMT---GAIHKQRRRRGSSSSVSSAEGAPPPRRDMRECYSCHERGHIARNC 275
>TC88387 similar to GP|13357253|gb|AAK20050.1 putative zinc finger protein
{Oryza sativa (japonica cultivar-group)}, partial (96%)
Length = 1286
Score = 28.5 bits (62), Expect = 2.4
Identities = 11/34 (32%), Positives = 15/34 (43%)
Frame = +1
Query: 157 RGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDC 190
RG GG + G C +C + GH +DC
Sbjct: 853 RGGGGSLRGGYRDGGFRDVVCRSCQQFGHMSRDC 954
Score = 28.5 bits (62), Expect = 2.4
Identities = 9/15 (60%), Positives = 10/15 (66%)
Frame = +1
Query: 177 CFNCHKMGHFKKDCP 191
C NC K GH +DCP
Sbjct: 730 CNNCRKTGHLARDCP 774
Score = 26.9 bits (58), Expect = 7.0
Identities = 7/17 (41%), Positives = 12/17 (70%)
Frame = +1
Query: 177 CFNCHKMGHFKKDCPEI 193
C NC + GH+ ++CP +
Sbjct: 424 CKNCKRPGHYVRECPNV 474
>TC89153 similar to GP|18855061|gb|AAL79753.1 putative RNA helicase {Oryza
sativa}, partial (3%)
Length = 737
Score = 28.5 bits (62), Expect = 2.4
Identities = 14/38 (36%), Positives = 18/38 (46%), Gaps = 3/38 (7%)
Frame = +3
Query: 157 RGNGGGRGNRRKSGNKSRF---ECFNCHKMGHFKKDCP 191
R +G NR S N+ CF+C + GH DCP
Sbjct: 78 RSSGYSSSNRSSSPNRRGSYGGACFSCGQPGHRASDCP 191
>TC80683 homologue to GP|8777424|dbj|BAA97014.1
gb|AAF56406.1~gene_id:K9P8.7~strong similarity to unknown
protein {Arabidopsis thaliana}, partial (16%)
Length = 1360
Score = 28.1 bits (61), Expect = 3.2
Identities = 8/16 (50%), Positives = 11/16 (68%)
Frame = +1
Query: 177 CFNCHKMGHFKKDCPE 192
C+ C K+GH +DC E
Sbjct: 1078 CYKCKKVGHLSRDCKE 1125
>TC91834 similar to PIR|T08416|T08416 disease resistance protein homolog
F18B3.230 - Arabidopsis thaliana, partial (3%)
Length = 803
Score = 28.1 bits (61), Expect = 3.2
Identities = 14/34 (41%), Positives = 24/34 (70%), Gaps = 1/34 (2%)
Frame = +2
Query: 66 MVESKAIMEQL-TEFNKILDDLENIEVQLEDEDK 98
+VE + ++ L ++FN I DDLE+I+ L+D D+
Sbjct: 107 VVEERTLVTGLESDFNDIKDDLESIQSFLKDADR 208
>TC81883 weakly similar to GP|7110148|gb|AAF36810.1| DNA
repair-recombination protein {Arabidopsis thaliana},
partial (10%)
Length = 1057
Score = 27.7 bits (60), Expect = 4.1
Identities = 26/94 (27%), Positives = 39/94 (40%), Gaps = 14/94 (14%)
Frame = +1
Query: 68 ESKAIMEQLTEFNKILDDLENIEVQL--------------EDEDKAILLLCALPKSFESF 113
E + + E+L ++ LDD+ I Q+ E D+ L + L K E
Sbjct: 334 ELQQVKEELDHKSQALDDVLGILAQVKTDKELVEPVVKYVEHADRIFLEIQTLQKKVEDL 513
Query: 114 KNTMLYGKEGTVTLEEIQAALRTKELTNSKDLTH 147
++ + G TLEEIQ L L +KD H
Sbjct: 514 ESELGCGGPEVRTLEEIQ--LELVALQGTKDNLH 609
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.132 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,731,075
Number of Sequences: 36976
Number of extensions: 81011
Number of successful extensions: 557
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 536
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 554
length of query: 240
length of database: 9,014,727
effective HSP length: 93
effective length of query: 147
effective length of database: 5,575,959
effective search space: 819665973
effective search space used: 819665973
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146972.3


