

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146972.1 + phase: 0 /pseudo
(594 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
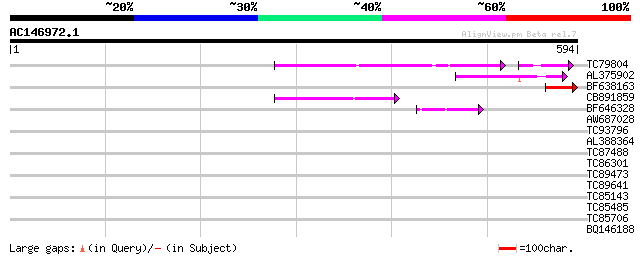
Score E
Sequences producing significant alignments: (bits) Value
TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like... 86 9e-21
AL375902 similar to GP|9759134|dbj mutator-like transposase-like... 70 3e-12
BF638163 68 9e-12
CB891859 similar to GP|6175165|gb| Mutator-like transposase {Ara... 59 4e-09
BF646328 similar to PIR|F84533|F845 Mutator-like transposase [im... 46 5e-05
AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~... 36 0.036
TC93796 similar to PIR|H71402|H71402 probable kinesin - Arabidop... 32 0.90
AL388364 similar to GP|5306274|gb|A expressed protein {Arabidops... 32 0.90
TC87488 30 3.4
TC86301 similar to GP|8809622|dbj|BAA97173.1 Myb-related transcr... 29 5.8
TC89473 similar to GP|9759049|dbj|BAB09571.1 gene_id:MVA3.7~pir|... 28 7.6
TC89641 similar to GP|9988432|dbj|BAB12698.1 hypothetical protei... 28 7.6
TC85143 similar to PIR|T11852|T11852 lipoxygenase (EC 1.13.11.12... 28 7.6
TC85485 similar to GP|22136900|gb|AAM91794.1 putative transketol... 28 7.6
TC85706 similar to GP|21700895|gb|AAM70571.1 At1g60730/F8A5_24 {... 28 9.9
BQ146188 similar to PIR|H96632|H966 hypothetical protein F8A5.26... 28 9.9
>TC79804 weakly similar to GP|6175165|gb|AAF04891.1| Mutator-like transposase
{Arabidopsis thaliana}, partial (57%)
Length = 2129
Score = 85.9 bits (211), Expect(2) = 9e-21
Identities = 58/242 (23%), Positives = 110/242 (44%)
Frame = +1
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMERVK 337
+G++ A++ P H +C+ HL +N K++KN L ++ A TT F M ++
Sbjct: 976 KGIVDAIRRKFPRSSHAFCMRHLSENIGKEFKNSRLIHLLWSAAYATTINAFREKMAEIE 1155
Query: 338 SINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIR 397
++ A +L + W YF Y +++ E FN IL + PI+ + E I+
Sbjct: 1156 EVSPNASMWLQHFHPSQWALVYFEGT--RYGHLSSNIEEFNKWILEAQELPIIQVIERIQ 1329
Query: 398 CYVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIASHNWTPLWNGDNARQRYHIENNCRI 457
+ + ++K L P + R+ + + + L + + + + + R
Sbjct: 1330 SKLKTEFDDRRLKSSSWCSVLTPSSERRMVEAINRASTYQVLKSDE---VEFEVISADRS 1500
Query: 458 KVDVDIFKQTCTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYFIQP 517
+ V+I +C+CR WQL +PC HA +AL + + T+ SYR+T + P
Sbjct: 1501 DI-VNIGSHSCSCRDWQLYGIPCSHAVAALISSRKDVYAYTAKCFTVASYRDTVCRGVTP 1677
Query: 518 MN 519
++
Sbjct: 1678 LS 1683
Score = 32.7 bits (73), Expect(2) = 9e-21
Identities = 16/57 (28%), Positives = 25/57 (43%)
Frame = +3
Query: 534 PPKVKRSAGRPKKNRRKDGNEEPIGGSSMKKTYNDIQCGRCGLMGHNSRSCAKQGVA 590
PPK +R GRP+K R I + + + C RC GH +C + ++
Sbjct: 1746 PPKFRRPPGRPEKKR--------ICVEDHNRDKHTVHCSRCNQTGHYKTTCKAEMIS 1892
>AL375902 similar to GP|9759134|dbj mutator-like transposase-like protein
{Arabidopsis thaliana}, partial (20%)
Length = 542
Score = 69.7 bits (169), Expect = 3e-12
Identities = 41/128 (32%), Positives = 54/128 (42%), Gaps = 11/128 (8%)
Frame = +2
Query: 468 CTCRFWQLT*MPCMHACSALALRGFKLEDHCHAWLTLGSYRETYSYFIQPMNSHIYWEPT 527
C+CR WQL PC HA +AL G T+ SYR YS I P+ W
Sbjct: 14 CSCRRWQLYGYPCAHAAAALISCGHNAHMFAEPCFTVQSYRMAYSQMIYPIPDKSQWREH 193
Query: 528 PYEKP-----------VPPKVKRSAGRPKKNRRKDGNEEPIGGSSMKKTYNDIQCGRCGL 576
PPK++R GRPKK + N K+ +QCGRC +
Sbjct: 194 GEGAEGGGGARVDIVIHPPKIRRPPGRPKKKVLRVEN--------FKRPKRVVQCGRCHM 349
Query: 577 MGHNSRSC 584
+GH+ + C
Sbjct: 350 LGHSQKKC 373
>BF638163
Length = 687
Score = 68.2 bits (165), Expect = 9e-12
Identities = 30/33 (90%), Positives = 31/33 (93%)
Frame = +2
Query: 562 MKKTYNDIQCGRCGLMGHNSRSCAKQGVARRPK 594
MKKTYND QCGRCGLMGH+SRSCAK GVARRPK
Sbjct: 380 MKKTYNDTQCGRCGLMGHSSRSCAK*GVARRPK 478
>CB891859 similar to GP|6175165|gb| Mutator-like transposase {Arabidopsis
thaliana}, partial (27%)
Length = 625
Score = 59.3 bits (142), Expect = 4e-09
Identities = 37/131 (28%), Positives = 64/131 (48%)
Frame = +1
Query: 278 QGLIPAMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARETTPTQFNRIMERVK 337
QG++ ++ P H +C+ HL + K++ N L ++ E A T +F + ++
Sbjct: 58 QGIVDGVEANFPTAFHGFCMRHLSDSFRKEFNNTMLVNLLWEAANCLTIIEFEGKVMEIE 237
Query: 338 SINQKAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIR 397
I+Q A ++ + P W AYF E + ++ N E NS IL G PI+ M E IR
Sbjct: 238 EISQDAAYWIRRVPPRLWATAYF-EGHRFGHLTANIVEALNSWILEASGLPIIQMMECIR 414
Query: 398 CYVMRKMSQNK 408
+M ++ +
Sbjct: 415 RQLMTWFNERR 447
>BF646328 similar to PIR|F84533|F845 Mutator-like transposase [imported] -
Arabidopsis thaliana, partial (3%)
Length = 636
Score = 45.8 bits (107), Expect = 5e-05
Identities = 22/70 (31%), Positives = 35/70 (49%)
Frame = -1
Query: 427 EKEKIASHNWTPLWNGDNARQRYHIENNCRIKVDVDIFKQTCTCRFWQLT*MPCMHACSA 486
EK+K + W W+GD +++ N K V++ ++TC CR W LT +PC H
Sbjct: 501 EKQK-SCRGWKATWHGDMEMNNFNVSNETN-KYIVNLAQRTCACRKWNLTGIPCAHVIPC 328
Query: 487 LALRGFKLED 496
+ G E+
Sbjct: 327 IWHNGLADEN 298
Score = 37.4 bits (85), Expect = 0.016
Identities = 20/52 (38%), Positives = 27/52 (51%)
Frame = -3
Query: 510 TYSYFIQPMNSHIYWEPTPYEKPVPPKVKRSAGRPKKNRRKDGNEEPIGGSS 561
TYS+ + P + W T E P +RSAGRPKK R+K +E G +
Sbjct: 172 TYSHIVLPSSGPKLWPVTNTEHINPLAKRRSAGRPKKLRKKANDETVRSGDA 17
>AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (11%)
Length = 662
Score = 36.2 bits (82), Expect = 0.036
Identities = 26/106 (24%), Positives = 37/106 (34%), Gaps = 4/106 (3%)
Frame = +1
Query: 283 AMQEVMPGVPHRYCVMHL*KNCTKQWKNKELRRVV*ECARET-TPTQFNRIMERVKSINQ 341
A+++V P HR C HL KN + K + TP QF + N+
Sbjct: 226 AIKQVFPDASHRLCAWHLHKNAQENIKKTPFLEGFRKAMYSNFTPEQFEDFWSELIQKNE 405
Query: 342 ---KAWEYLNKWPKEAWIKAYFNENCKAYNIVNNACEVFNSKILNY 384
AW K W AY + + CE N+ + Y
Sbjct: 406 LEGNAWVIKTYANKSLWATAYLRDKFFGRIRTTSQCEAINAIVKTY 543
>TC93796 similar to PIR|H71402|H71402 probable kinesin - Arabidopsis
thaliana, partial (5%)
Length = 744
Score = 31.6 bits (70), Expect = 0.90
Identities = 9/25 (36%), Positives = 15/25 (60%)
Frame = +2
Query: 468 CTCRFWQLT*MPCMHACSALALRGF 492
C CR+W++ + C+H C +GF
Sbjct: 647 CHCRYWRVVGINCLHVCGNFGYQGF 721
>AL388364 similar to GP|5306274|gb|A expressed protein {Arabidopsis
thaliana}, partial (4%)
Length = 524
Score = 31.6 bits (70), Expect = 0.90
Identities = 21/62 (33%), Positives = 29/62 (45%), Gaps = 6/62 (9%)
Frame = -2
Query: 325 TPTQFNRIMERVKSINQKAWEYLNKWPKE---AWIKAYFNENCKAYN---IVNNACEVFN 378
T +F RI + + N YLNKWP++ +I F N KAY I + C + N
Sbjct: 307 TAAKFRRIKQLLLIHNALGL*YLNKWPQDVQSVYISVVFTLNDKAYRYLLITSFCCTLQN 128
Query: 379 SK 380
K
Sbjct: 127 CK 122
>TC87488
Length = 1591
Score = 29.6 bits (65), Expect = 3.4
Identities = 16/44 (36%), Positives = 25/44 (56%), Gaps = 2/44 (4%)
Frame = +3
Query: 421 WQQSRLEKEKIASHN--WTPLWNGDNARQRYHIENNCRIKVDVD 462
W++ K+ I +N W P++NG+N Y I N+ I +DVD
Sbjct: 699 WKEIATRKQVILKYNFLWKPVYNGEN--HIYWITNDGVIVMDVD 824
>TC86301 similar to GP|8809622|dbj|BAA97173.1 Myb-related transcription
activator-like {Arabidopsis thaliana}, partial (56%)
Length = 1396
Score = 28.9 bits (63), Expect = 5.8
Identities = 10/20 (50%), Positives = 13/20 (65%)
Frame = +2
Query: 570 QCGRCGLMGHNSRSCAKQGV 589
+C C GHNSR+C +GV
Sbjct: 194 RCSHCSHNGHNSRTCPNRGV 253
>TC89473 similar to GP|9759049|dbj|BAB09571.1
gene_id:MVA3.7~pir||T13462~similar to unknown protein
{Arabidopsis thaliana}, partial (5%)
Length = 715
Score = 28.5 bits (62), Expect = 7.6
Identities = 17/67 (25%), Positives = 32/67 (47%), Gaps = 2/67 (2%)
Frame = +2
Query: 361 NENCKAYNIVNNACEVFNSKILNYRGHPILTMAEEIRCYVMRKMSQNKMKLDGRAGPLC- 419
+ +C +NN VFNS I+ Y I + + + C + R + + ++DG A C
Sbjct: 119 SSSCSPKKWINNCIRVFNS-IICYIVFLIFDLLDAVLCVIYRYLDE---RIDGVASSCCC 286
Query: 420 -PWQQSR 425
W++ +
Sbjct: 287 SKWERQK 307
>TC89641 similar to GP|9988432|dbj|BAB12698.1 hypothetical protein {Oryza
sativa (japonica cultivar-group)}, partial (44%)
Length = 1080
Score = 28.5 bits (62), Expect = 7.6
Identities = 9/15 (60%), Positives = 12/15 (80%)
Frame = +3
Query: 570 QCGRCGLMGHNSRSC 584
+C CG +GHNSR+C
Sbjct: 93 KCSHCGNIGHNSRTC 137
>TC85143 similar to PIR|T11852|T11852 lipoxygenase (EC 1.13.11.12) - kidney
bean, partial (93%)
Length = 3009
Score = 28.5 bits (62), Expect = 7.6
Identities = 17/72 (23%), Positives = 31/72 (42%), Gaps = 2/72 (2%)
Frame = +3
Query: 398 CYVMRKMSQNKMKLDGRAGPLCPWQQSRLEKEKIAS--HNWTPLWNGDNARQRYHIENNC 455
C M + + +A C W+ + ++ H WT G++ R+ H
Sbjct: 1155 CKSMCDSKTSSVSTRKQARSQCLWRSN*YNNKRTLGD*HGWTHSRKGNSRREAIHFGLP* 1334
Query: 456 RIKVDVDIFKQT 467
RI V ++++KQT
Sbjct: 1335 RIHVILEVYKQT 1370
>TC85485 similar to GP|22136900|gb|AAM91794.1 putative transketolase
{Arabidopsis thaliana}, partial (90%)
Length = 2728
Score = 28.5 bits (62), Expect = 7.6
Identities = 9/27 (33%), Positives = 18/27 (66%)
Frame = -1
Query: 193 IWRTRTTRSGFWNYSMQILEIMCVMDG 219
+WRT++ SG+ N S + + +C ++G
Sbjct: 1168 VWRTKSNGSGYLNQSRFVCDSLCFLNG 1088
>TC85706 similar to GP|21700895|gb|AAM70571.1 At1g60730/F8A5_24 {Arabidopsis
thaliana}, partial (96%)
Length = 1416
Score = 28.1 bits (61), Expect = 9.9
Identities = 9/17 (52%), Positives = 14/17 (81%)
Frame = +1
Query: 227 RYEFPFKWMELYYLLIY 243
+YEFPF+W+ LY ++Y
Sbjct: 1174 QYEFPFEWIWLYKTMLY 1224
>BQ146188 similar to PIR|H96632|H966 hypothetical protein F8A5.26 [imported]
- Arabidopsis thaliana, partial (12%)
Length = 397
Score = 28.1 bits (61), Expect = 9.9
Identities = 9/17 (52%), Positives = 14/17 (81%)
Frame = +2
Query: 227 RYEFPFKWMELYYLLIY 243
+YEFPF+W+ LY ++Y
Sbjct: 251 QYEFPFEWIWLYKTMLY 301
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.350 0.154 0.574
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,383,336
Number of Sequences: 36976
Number of extensions: 380463
Number of successful extensions: 4032
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 2212
Number of HSP's successfully gapped in prelim test: 227
Number of HSP's that attempted gapping in prelim test: 1719
Number of HSP's gapped (non-prelim): 2595
length of query: 594
length of database: 9,014,727
effective HSP length: 102
effective length of query: 492
effective length of database: 5,243,175
effective search space: 2579642100
effective search space used: 2579642100
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146972.1


