

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146941.15 - phase: 0
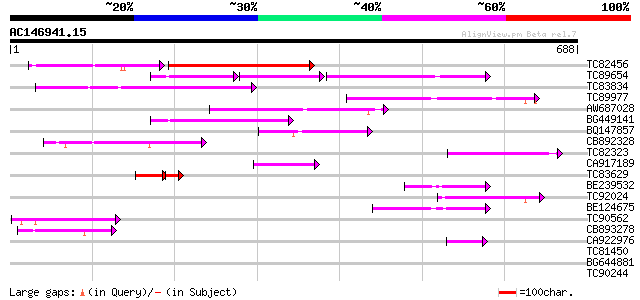
(688 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far... 227 1e-72
TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein... 123 2e-68
TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabid... 186 3e-47
TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired re... 135 4e-32
AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~... 134 9e-32
BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidop... 125 7e-29
BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {A... 82 9e-16
CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sin... 76 5e-14
TC82323 similar to GP|7673677|gb|AAF66982.1| transposase {Zea ma... 65 7e-11
CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.3... 62 7e-10
TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNa... 45 8e-10
BE239532 58 1e-08
TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10... 57 2e-08
BE124675 weakly similar to GP|13365573|db hypothetical protein~s... 55 7e-08
TC90562 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20... 52 6e-07
CB893278 weakly similar to PIR|T05645|T056 hypothetical protein ... 52 1e-06
CA922976 weakly similar to GP|9502168|gb|A contains simlarity to... 45 9e-05
TC81450 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20... 39 0.005
BG644881 similar to GP|6648604|gb| ubiquitin-specific protease; ... 33 0.36
TC90244 similar to GP|15450749|gb|AAK96646.1 At2g39750/T5I7.5 {A... 29 5.3
>TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far-red
impaired response protein {Oryza sativa}, partial (7%)
Length = 1244
Score = 227 bits (579), Expect(2) = 1e-72
Identities = 98/177 (55%), Positives = 134/177 (75%)
Frame = +2
Query: 193 GDGKALMSYFSKMREQNSNFFYDIDLDDDFHVRNVFWADARSRATYEYFGDVVTFDTTYL 252
GD A+ S+F +M++QNS F+ +D+DD +++N+FWADAR RA YEYFG+V+T DTTYL
Sbjct: 713 GDVDAIHSFFHRMQKQNSQFYCAMDMDDKRNIQNLFWADARCRAAYEYFGEVITLDTTYL 892
Query: 253 TNKFDMPFATFVGVNHHGQSTLLGCGLLSGEDTESFVWLFKSWLRCMLGKAPVGIVTDQC 312
T+K+D+P FVGVNHHGQ+ LLGC +LS D ++ WLF WL CM G AP GI+T++
Sbjct: 893 TSKYDLPLVPFVGVNHHGQTILLGCAILSNLDAKTLTWLFTRWLECMHGHAPNGIITEED 1072
Query: 313 KAVQNAIELVFPTTRHRWCLWHIMKKIPEKLNEYSEYKRIKSAMEGAVYDTHTTTGF 369
KA++NAIE+ FP RHRWCLWHIMKK+PE L +YS + IK+ ++ VYD+ + + F
Sbjct: 1073KAMKNAIEVAFPKARHRWCLWHIMKKVPEMLGKYSRNESIKTLLQDVVYDSMSKSDF 1243
Score = 65.1 bits (157), Expect(2) = 1e-72
Identities = 52/175 (29%), Positives = 83/175 (46%), Gaps = 9/175 (5%)
Frame = +1
Query: 23 SDDDEACWKPATGMRFSCIDDVKTLYREYALKKGFRWKTRTSKKRDDGETCYAILV-CTR 81
S+D+E P TGM F ++V Y YA + GF +SK DDG+ + + C R
Sbjct: 193 SEDEE----PRTGMTFRSEEEVIRYYMNYANRVGFGVTKISSKNADDGKKYFTLACNCAR 360
Query: 82 EGSQGSEIPCTLKTPPSKTKNCPAKICIKLEKDGLWYISKFESRHSHKTSPTK----RMD 137
S+ P + + SKT+ C A++ + DG IS+ H+H+ TK R+D
Sbjct: 361 RYVSTSKNP-SKQYLTSKTQ-CRARLNACVALDGSSTISRIVLEHNHELGQTKGRYFRLD 534
Query: 138 ----LHVKRTIEINEDAGVRTIKTFRSIVNNAEGHENIPFCEKDMINYVNNEQHL 188
HV+R +E+N+ G+ + +S+ G N+ E D N+V + L
Sbjct: 535 ESSGTHVERNLELNDQGGINVDRNLQSVGLEMNGCGNLTSGENDCRNFVQKVRRL 699
>TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein F20D10.290
- Arabidopsis thaliana, partial (65%)
Length = 1358
Score = 123 bits (308), Expect(3) = 2e-68
Identities = 63/199 (31%), Positives = 100/199 (49%)
Frame = +1
Query: 385 NDWLSGLYEERHRWAPTFVRKYFWAGMSTTQRSESMHAFFDGYINSTTSLNQFVKQYDNA 444
N+W +Y R W P ++R F+ + +E ++ FFDG++N++T+L V+QY+ A
Sbjct: 697 NEWPQSMYNARQHWVPVYLRGSFFGEIPLNDGNECLNFFFDGHVNASTTLQLLVRQYEKA 876
Query: 445 LSSRAEKEFEADFNSLDTTIPCVSNSSIEKQLQGEYTHAKFKEVQAEFISKMNCAPSLNV 504
+S+ E+E +ADF + ++ + S +EKQ YT F + Q E + M +
Sbjct: 877 VSTWHERELKADFETSNSNPVLRTPSPMEKQAASLYTRKMFMKFQEELVGTMANPATKIE 1056
Query: 505 VEGCFATYHVLEEVVVGGRHKENVFKVVFNQENQDFSCECSLFEFRGILCCHVLSVCTRE 564
G TY V + + V FN SC C +FE+ GI+C H+L+V +
Sbjct: 1057DSGTITTYRVAK-----FGENQTSHTVAFNSSEMKASCSCQMFEYSGIVCRHILAVFRAK 1221
Query: 565 RVKNVPEKYVLTRWKKNIK 583
V +P YVL RW N K
Sbjct: 1222NVLTLPSHYVLKRWTMNAK 1278
Score = 100 bits (249), Expect(3) = 2e-68
Identities = 48/106 (45%), Positives = 61/106 (57%)
Frame = +2
Query: 172 PFCEKDMINYVNNEQHLIGKEGDGKALMSYFSKMREQNSNFFYDIDLDDDFHVRNVFWAD 231
P C K + + Q +G G G ++ Y +MR +N FFY + D D N+FWAD
Sbjct: 59 PNCCKL*AHMSSTRQRTLG--GGGHQVLDYLKRMRAENHAFFYAVQSDVDNAGGNIFWAD 232
Query: 232 ARSRATYEYFGDVVTFDTTYLTNKFDMPFATFVGVNHHGQSTLLGC 277
R Y YFGD V FDTTY TN++ +PFA+F G NHHGQ L GC
Sbjct: 233 ETCRTNYSYFGDTVIFDTTYKTNQYRVPFASFTGFNHHGQPVLFGC 370
Score = 75.9 bits (185), Expect(3) = 2e-68
Identities = 36/105 (34%), Positives = 58/105 (54%), Gaps = 1/105 (0%)
Frame = +3
Query: 279 LLSGEDTESFVWLFKSWLRCMLGKAPVGIVTDQCKAVQNAIELVFPTTRHRWCLWHIMKK 338
L+ E S++WLFK+WLR + G+ PV I TD +Q A+ V P TRHR+C W I ++
Sbjct: 375 LILNESEPSYIWLFKTWLRAVSGRPPVSITTDLDPVIQVAVAQVLPPTRHRFCKWSIFRE 554
Query: 339 IPEKL-NEYSEYKRIKSAMEGAVYDTHTTTGFEEKWCSFIDKFML 382
KL + Y + + V+++ T FE W S ++++ +
Sbjct: 555 NRSKLAHLYQSNPTFDNEFKKCVHESVTIDEFESCWRSLLERYYI 689
>TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabidopsis
thaliana, partial (24%)
Length = 1032
Score = 186 bits (472), Expect = 3e-47
Identities = 98/269 (36%), Positives = 154/269 (56%), Gaps = 1/269 (0%)
Frame = +3
Query: 32 PATGMRFSCIDDVKTLYREYALKKGFRWKTRTSK-KRDDGETCYAILVCTREGSQGSEIP 90
PA M F DD + Y YA + GF + + S KR+ E A+L C+ +G + ++
Sbjct: 213 PALAMEFESYDDAYSYYICYAKEVGFCVRVKNSWFKRNSKEKYGAVLCCSSQGFKRTKDV 392
Query: 91 CTLKTPPSKTKNCPAKICIKLEKDGLWYISKFESRHSHKTSPTKRMDLHVKRTIEINEDA 150
L+ CPA I +KL + W I + H+H ++ +K+ + DA
Sbjct: 393 NNLRK--ETRTGCPAMIRMKLVESQRWRICEVTLEHNHVLGA--KIHKSIKKNSLPSSDA 560
Query: 151 GVRTIKTFRSIVNNAEGHENIPFCEKDMINYVNNEQHLIGKEGDGKALMSYFSKMREQNS 210
+TIK + ++V + EG++N+ +D + L ++GD +A+ ++ +M+ N
Sbjct: 561 EGKTIKVYHALVIDTEGNDNLNSNARDDRAFSKYSNKLNLRKGDTQAIYNFLCRMQLTNP 740
Query: 211 NFFYDIDLDDDFHVRNVFWADARSRATYEYFGDVVTFDTTYLTNKFDMPFATFVGVNHHG 270
NFFY +D +D+ H+RN W DA+SRA YF DV+ FD TYL NK+++P VG+NHHG
Sbjct: 741 NFFYLMDFNDEGHLRNALWVDAKSRAACGYFSDVIYFDNTYLVNKYEIPLVALVGINHHG 920
Query: 271 QSTLLGCGLLSGEDTESFVWLFKSWLRCM 299
QS LLGCGLL+GE ES+ WLF++W++C+
Sbjct: 921 QSVLLGCGLLAGEIIESYKWLFRTWIKCI 1007
>TC89977 similar to GP|5764395|gb|AAD51282.1| far-red impaired response
protein {Arabidopsis thaliana}, partial (35%)
Length = 1523
Score = 135 bits (341), Expect = 4e-32
Identities = 85/242 (35%), Positives = 128/242 (52%), Gaps = 8/242 (3%)
Frame = +2
Query: 409 AGMSTTQRSESMHAFFDGYINSTTSLNQFVKQYDNALSSRAEKEFEADFNSLDTTIPCVS 468
AG ST QRSESM++FFD YI+ +L +FVKQY L +R ++E ADF++L S
Sbjct: 2 AGTSTAQRSESMNSFFDKYIHKKITLKEFVKQYRLILLNRYDEEEIADFDTLHKQPALKS 181
Query: 469 NSSIEKQLQGEYTHAKFKEVQAEFISKMNCAPSLNVVEGCFATYHVLEEVVVGGRHKENV 528
S EKQ+ YTHA FK+ Q E + C + V +G Y +V K+
Sbjct: 182 PSPWEKQMSTIYTHAIFKKFQVEVLGVAGCQSRIEVGDGTAVRY------IVQDYEKDEE 343
Query: 529 FKVVFNQENQDFSCECSLFEFRGILCCHVLSVCTRERVKNVPEKYVLTRWKKNIKRKHSY 588
F V + + + + SC C LFE++G LC H LSV R +VP Y++ RW K+ K +
Sbjct: 344 FLVTWKELSSEVSCFCRLFEYKGFLCRHALSVLQRCGCSSVPSHYIMKRWTKDAKIRE-- 517
Query: 589 IKSSYCARELKPKMDRFDKLCKHFYEIAEVAAESED----ATKALHETLHQF----NSNK 640
++ + ++ + R++ LCK ++E + SE+ A +AL + L NSN
Sbjct: 518 -PAADRTQRIQTRPQRYNDLCKRSIALSEEGSSSEESYNIALRALIDALKNCVLVNNSND 694
Query: 641 DG 642
+G
Sbjct: 695 NG 700
>AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (11%)
Length = 662
Score = 134 bits (338), Expect = 9e-32
Identities = 76/225 (33%), Positives = 108/225 (47%), Gaps = 8/225 (3%)
Frame = +1
Query: 243 DVVTFDTTYLTNKFDMPFATFVGVNHHGQSTLLGCGLLSGEDTESFVWLFKSWLRCMLGK 302
DV+ FDTTY NK++ P F G NHH Q+ + G LL E ES+ W+ +L CM K
Sbjct: 1 DVLAFDTTYKKNKYNYPLCIFSGCNHHSQTIIFGVALLEDETIESYKWVLNRFLECMENK 180
Query: 303 APVGIVTDQCKAVQNAIELVFPTTRHRWCLWHIMKKIPEKLNEYSEYKRIKSAMEGAVYD 362
P +VTD +++ AI+ VFP HR C WH+ K E + + + + AM Y
Sbjct: 181 FPKAVVTDGDGSMREAIKQVFPDASHRLCAWHLHKNAQENIKKTPFLEGFRKAM----YS 348
Query: 363 THTTTGFEEKWCSFIDKFMLQQNDWLSGLYEERHRWAPTFVRKYFWAGMSTTQRSESMHA 422
T FE+ W I K L+ N W+ Y + WA ++R F+ + TT + E+++A
Sbjct: 349 NFTPEQFEDFWSELIQKNELEGNAWVIKTYANKSLWATAYLRDKFFGRIRTTSQCEAINA 528
Query: 423 FFDGYINSTTS--------LNQFVKQYDNALSSRAEKEFEADFNS 459
Y S LN+F + Y N E ADF S
Sbjct: 529 IVKTYSRSQRQNF*IYA*FLNRF*EGYRN-------NELVADFKS 642
>BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (33%)
Length = 690
Score = 125 bits (313), Expect = 7e-29
Identities = 63/174 (36%), Positives = 99/174 (56%)
Frame = +3
Query: 171 IPFCEKDMINYVNNEQHLIGKEGDGKALMSYFSKMREQNSNFFYDIDLDDDFHVRNVFWA 230
+PF EKD+ N + + + L +E + L+ +++++ NF ++ LD + + N+ W+
Sbjct: 165 LPFTEKDVRNLLQSFRKLDPEE-ETLDLLRMCRNIKDKDPNFKFEYTLDANNRLENIAWS 341
Query: 231 DARSRATYEYFGDVVTFDTTYLTNKFDMPFATFVGVNHHGQSTLLGCGLLSGEDTESFVW 290
A S Y+ FGD V FDTT+ FDMP +VG+N++G GC LL E SF W
Sbjct: 342 YASSIQLYDIFGDAVVFDTTHRLTAFDMPLGLWVGINNYGMPCFFGCVLLRDETVRSFSW 521
Query: 291 LFKSWLRCMLGKAPVGIVTDQCKAVQNAIELVFPTTRHRWCLWHIMKKIPEKLN 344
K++L M GKAP I+TDQ ++ A+ P T+H +C+W I+ K P +N
Sbjct: 522 AIKAFLGFMNGKAPQTILTDQNICLKEALSAEMPMTKHAFCIWMIVAKFPSWVN 683
>BQ147857 weakly similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (26%)
Length = 560
Score = 81.6 bits (200), Expect = 9e-16
Identities = 41/143 (28%), Positives = 79/143 (54%), Gaps = 5/143 (3%)
Frame = +1
Query: 303 APVGIVTDQCKAVQNAIELVFPTTRHRWCLWHIMKKIPEKL-----NEYSEYKRIKSAME 357
AP ++TD ++ AI + P ++H +C+WHI+ K + ++Y E+K A
Sbjct: 1 APQTLLTDHNTWLKEAIAVEMPESKHAFCIWHILSKFSDWXYLLLGSQYDEWK----ADF 168
Query: 358 GAVYDTHTTTGFEEKWCSFIDKFMLQQNDWLSGLYEERHRWAPTFVRKYFWAGMSTTQRS 417
+Y+ FE+ W +DK+ L N + LY R WA F+R+YF+AG+++ ++
Sbjct: 169 HRLYNLEMXEDFEKSWRQMVDKYGLHANKHIISLYSLRTFWALPFLRRYFFAGLTSXCQT 348
Query: 418 ESMHAFFDGYINSTTSLNQFVKQ 440
ES++ F ++++ + +F++Q
Sbjct: 349 ESINVFIQRFLSAQSQPXRFLQQ 417
>CB892328 GP|15075873|em PROBABLE GLYCOSYL HYDROLASE PROTEIN {Sinorhizobium
meliloti}, partial (1%)
Length = 828
Score = 75.9 bits (185), Expect = 5e-14
Identities = 55/212 (25%), Positives = 100/212 (46%), Gaps = 15/212 (7%)
Frame = +1
Query: 42 DDVKTLYREYALKKGFRWKTRTSKK----RDDGETCYAILVCTREGSQGSEIP--CTLKT 95
D+ LY+E+A K GF R K+ + +T C+++G + +E K
Sbjct: 184 DEAYDLYQEHAFKTGF--SVRKGKELYYDNEKKKTRLKDFYCSKQGFKNNEPDGEVAYKR 357
Query: 96 PPSKTKNCPAKICIKLEKDGLWYISKFESRHSHKTSPTK-RMDLHVKRTIEINEDAGVRT 154
S+T NC A + + K+G+W ++K H+H+ P + R L R + ++ +++
Sbjct: 358 ADSRT-NCLAMVRFNVTKEGVWKVTKLILDHNHEFVPLQQRYLLRSMRNMSNLKEDRIKS 534
Query: 155 IKTFRSIVNNAEGH--------ENIPFCEKDMINYVNNEQHLIGKEGDGKALMSYFSKMR 206
+ V N G+ + KDM NYV+ + + GD ++L+++ +
Sbjct: 535 LVNDAIRVTNVGGYLGKEVGGVDKRGVMLKDMHNYVSTGKLKFIEAGDAQSLLNHLQSKQ 714
Query: 207 EQNSNFFYDIDLDDDFHVRNVFWADARSRATY 238
Q+S F+Y + LD + + NVFW D +S Y
Sbjct: 715 AQDSMFYYSVQLDHESRLNNVFWRDGKSVVDY 810
>TC82323 similar to GP|7673677|gb|AAF66982.1| transposase {Zea mays},
partial (2%)
Length = 1165
Score = 65.5 bits (158), Expect = 7e-11
Identities = 41/140 (29%), Positives = 63/140 (44%), Gaps = 1/140 (0%)
Frame = +1
Query: 532 VFNQENQDFSCECSLFEFRGILCCHVLSVCTRERVKNVPEKYVLTRWKKNIKRKHSYIKS 591
V+N + +F C C LFE RGI CCHV V E V ++P+ V+TRW KN K + S
Sbjct: 1 VYNAASLEFQCSCRLFESRGIPCCHVFFVMKEEDVDHIPKCLVMTRWTKNAKGEFLNSDS 180
Query: 592 SYCARELKPKMDRFDKLCKHFYEIAEVAAESEDATKALHETLHQFNSNKDGITDIV-NRS 650
+ ++ RF C F + A++ + + + + + D ++S
Sbjct: 181 NGEIDANMIELARFGAYCSAFTTFCKEASKKNGVFRDIMDEILNLQKKYCDVEDSTRSKS 360
Query: 651 FIDDSNPNNGIGIHSPVRVK 670
F DD + PV VK
Sbjct: 361 FTDDQ-------VGDPVTVK 399
>CA917189 similar to PIR|T05645|T05 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (12%)
Length = 312
Score = 62.0 bits (149), Expect = 7e-10
Identities = 31/82 (37%), Positives = 44/82 (52%), Gaps = 1/82 (1%)
Frame = +1
Query: 296 LRCMLGKAPVGIVTDQCKAVQNAIELVFPTTRHRWCLWHIMKKIPEKLNE-YSEYKRIKS 354
L M G+ P+ I TD +Q+AI VFP TRHR+C WHI K+ EKL+ + ++ ++
Sbjct: 1 LTAMSGRPPLSITTDHDSVIQSAIMQVFPDTRHRFCKWHIFKQCQEKLSHIFLQFPNFEA 180
Query: 355 AMEGAVYDTHTTTGFEEKWCSF 376
V T + FE W F
Sbjct: 181 EFHKCVNLTDSIDEFESCWSHF 246
>TC83629 weakly similar to GP|6692261|gb|AAF24611.1| putative RNase H
{Arabidopsis thaliana}, partial (21%)
Length = 1071
Score = 45.4 bits (106), Expect(2) = 8e-10
Identities = 20/39 (51%), Positives = 26/39 (66%)
Frame = +2
Query: 153 RTIKTFRSIVNNAEGHENIPFCEKDMINYVNNEQHLIGK 191
R KTF+ +V + HE PFC++DM NYVN E+H I K
Sbjct: 2 RIKKTFQPLVKDVGAHEITPFCKRDMGNYVNKERHAIWK 118
Score = 36.2 bits (82), Expect(2) = 8e-10
Identities = 15/22 (68%), Positives = 17/22 (77%)
Frame = +3
Query: 190 GKEGDGKALMSYFSKMREQNSN 211
GKEGDG L+SYF MR+QN N
Sbjct: 114 GKEGDGMVLISYFCNMRKQNQN 179
>BE239532
Length = 645
Score = 57.8 bits (138), Expect = 1e-08
Identities = 31/105 (29%), Positives = 52/105 (49%), Gaps = 1/105 (0%)
Frame = +3
Query: 480 YTHAKFKEVQAEFISKMNCAPSLNVVEGCFATYHVLEEVVVGGRHKENV-FKVVFNQENQ 538
YTH F+ E++ + S+ + + + H EV++ HK N + V F+ N
Sbjct: 66 YTHKGFRIFLNEYLEGTGGSTSIEISQSNNLSNH---EVMLN--HKPNKKYAVTFDSSNM 230
Query: 539 DFSCECSLFEFRGILCCHVLSVCTRERVKNVPEKYVLTRWKKNIK 583
+C C F+ GILC H L + + + +P++Y L RW KN +
Sbjct: 231 KINCSCRKFDSMGILCSHALRIYNIKGILRIPDQYFLKRWSKNAR 365
>TC92024 similar to PIR|T05645|T05645 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (11%)
Length = 938
Score = 57.4 bits (137), Expect = 2e-08
Identities = 43/137 (31%), Positives = 62/137 (44%), Gaps = 8/137 (5%)
Frame = +3
Query: 520 VGGRHKENVFKVVFNQENQDFSCECSLFEFRGILCCHVLSVCTRERVKNVPEKYVLTRWK 579
+G HK + V FN +C C +FEF G+LC H+L+V V +P Y+L RW
Sbjct: 33 LGEDHK--AYNVRFNVLEMKATCSCQMFEFSGLLCRHILAVFRVTNVLTLPSHYILKRWT 206
Query: 580 KNIKRKHSYIKSSYCARE--LKPKMDRFDKLCKHFYEIAEVAAESED----ATKALHETL 633
KN K S + S A L+ R++ L ++ + A S + A AL E
Sbjct: 207 KNAKSNVSLQEHSSHAYTYYLESHTVRYNTLRHEAFKFVDKGASSPETYDVAKDALQEAA 386
Query: 634 HQFNS--NKDGITDIVN 648
+ K+G T I N
Sbjct: 387 KRVAQVMRKEGRTPISN 437
>BE124675 weakly similar to GP|13365573|db hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (4%)
Length = 448
Score = 55.5 bits (132), Expect = 7e-08
Identities = 35/143 (24%), Positives = 58/143 (40%)
Frame = +2
Query: 441 YDNALSSRAEKEFEADFNSLDTTIPCVSNSSIEKQLQGEYTHAKFKEVQAEFISKMNCAP 500
++ + + KE EA + N + K YT F+ Q + +N
Sbjct: 2 FERIVEEQRYKEIEASDEMKGCLPKLMGNVVVLKHASVAYTPRAFEVFQQRYEKSLNVIV 181
Query: 501 SLNVVEGCFATYHVLEEVVVGGRHKENVFKVVFNQENQDFSCECSLFEFRGILCCHVLSV 560
+ + +G Y +V G ++ + V F+ + C C FE G LC H L V
Sbjct: 182 NQHKRDGYLFEY----KVNTYGHARQ--YTVTFSSSDNTVVCSCMKFEHVGFLCSHALKV 343
Query: 561 CTRERVKNVPEKYVLTRWKKNIK 583
+K VP +Y+L RW K+ +
Sbjct: 344 LDNRNIKVVPSRYILKRWTKDAR 412
>TC90562 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20
{Arabidopsis thaliana}, partial (66%)
Length = 1169
Score = 52.4 bits (124), Expect = 6e-07
Identities = 39/153 (25%), Positives = 66/153 (42%), Gaps = 21/153 (13%)
Frame = +2
Query: 3 SSENLVDCGSI-------HDEGMEVDNSDDDEACW------------KPATGMRFSCIDD 43
+++ +V+C S D+ M D+S D+ +P G F +
Sbjct: 179 ANKEIVECSSFDPTDEQDQDDTMTQDSSGVDQIASAVPVTSVMRTIDEPYIGQEFVSEAE 358
Query: 44 VKTLYREYALKKGFRWK-TRTSKKRDDGETCYAILVCTREGSQGSEIPCTLKTPPSKTK- 101
Y YA + GF + ++ S+ R DG LVC +EG + + + ++T+
Sbjct: 359 AHAFYNSYATRVGFVIRVSKLSRSRRDGSVIGRALVCNKEGFRMPDKREKIVRQRAETRV 538
Query: 102 NCPAKICIKLEKDGLWYISKFESRHSHKTSPTK 134
C A I ++ GLW I+KF H+H +P K
Sbjct: 539 GCRAMIMVRKLNSGLWSITKFVKEHTHPLTPGK 637
>CB893278 weakly similar to PIR|T05645|T056 hypothetical protein F20D10.300 -
Arabidopsis thaliana, partial (5%)
Length = 821
Score = 51.6 bits (122), Expect = 1e-06
Identities = 33/125 (26%), Positives = 58/125 (46%), Gaps = 5/125 (4%)
Frame = +3
Query: 10 CGSIHDEGMEVDNSDDDEACWKPATGMRFSCIDDVKTLYREYALKKGFRWKTRTSKK-RD 68
C E +E +D D +P GM+F+ ++ + Y Y + GF + +++ R
Sbjct: 255 CTGEDKEFVEQPVADSDTL--EPFIGMQFNSREEARGFYDGYGRRIGFTVRIHHNRRSRV 428
Query: 69 DGETCYAILVCTREGSQGSEI----PCTLKTPPSKTKNCPAKICIKLEKDGLWYISKFES 124
+ E VC++EG + + L PP+ + C A I + L +G W ++KF
Sbjct: 429 NNELIGQDFVCSKEGFRAKKYVHRKDRVLPPPPATREGCQAMIRLALRDEGKWVVTKFVK 608
Query: 125 RHSHK 129
H+HK
Sbjct: 609 EHTHK 623
>CA922976 weakly similar to GP|9502168|gb|A contains simlarity to Arabidopsis
thaliana far-red impaired response protein
(GB:AAD51282.1), partial (11%)
Length = 827
Score = 45.1 bits (105), Expect = 9e-05
Identities = 19/50 (38%), Positives = 27/50 (54%)
Frame = -3
Query: 531 VVFNQENQDFSCECSLFEFRGILCCHVLSVCTRERVKNVPEKYVLTRWKK 580
V++ E + C C FE GILC H L + + +PEKY L+RW +
Sbjct: 735 VLWLPEKEQILCSCKEFESSGILCRHALRILVTKNYFQLPEKYYLSRWHR 586
>TC81450 similar to GP|16604382|gb|AAL24197.1 AT3g59470/T16L24_20
{Arabidopsis thaliana}, partial (59%)
Length = 832
Score = 39.3 bits (90), Expect = 0.005
Identities = 23/80 (28%), Positives = 40/80 (49%), Gaps = 2/80 (2%)
Frame = +2
Query: 61 TRTSKKRDDGETCYAILVCTREGSQGSEIPCTLKTPPSKTK-NCPAKICIKLEKDGLWYI 119
++ S+ R DG LVC +EG + + + ++T+ C A I ++ G W I
Sbjct: 128 SKLSRSRKDGTAIGRALVCNKEGYRMPDKREKIVRQRAETRVGCRAMIMMRKINSGKWVI 307
Query: 120 SKFESRHSHKTSP-TKRMDL 138
+KF H+H +P + R D+
Sbjct: 308 TKFVKEHTHPLNPGSSRRDM 367
>BG644881 similar to GP|6648604|gb| ubiquitin-specific protease; UBP5
{Arabidopsis thaliana}, partial (20%)
Length = 767
Score = 33.1 bits (74), Expect = 0.36
Identities = 35/166 (21%), Positives = 69/166 (41%), Gaps = 10/166 (6%)
Frame = -3
Query: 111 LEKDGLWYISKFESRHSHKTSPTKRMDL-HVKRTIEINEDAGVRTIKTFRSIVNNAEGHE 169
L + +WY K + R +K++DL + + I+ ++ RS+ + E
Sbjct: 537 LVPEDMWYCPKCKERRQ----ASKKLDLWRLPEVLVIH----LKRFSYSRSMKHKLETFV 382
Query: 170 NIPFCEKDMINYVNNEQHLIGKEGDGKALMSYFSKM---------REQNSNFFYDIDLDD 220
N P + D+ NY+ N+ + + + AL +++ M + + N +Y+ D
Sbjct: 381 NFPIHDFDLTNYIANKNNSRRQVYELYALTNHYGSMGSGHYTAHIKLLDENRWYNF---D 211
Query: 221 DFHVRNVFWADARSRATYEYFGDVVTFDTTYLTNKFDMPFATFVGV 266
D H+ + D + A Y F V D ++N F+G+
Sbjct: 210 DSHISLISEDDVNTAAAYVLFYRRVKTDDDIVSNGV*SCMHAFLGI 73
>TC90244 similar to GP|15450749|gb|AAK96646.1 At2g39750/T5I7.5 {Arabidopsis
thaliana}, partial (49%)
Length = 1319
Score = 29.3 bits (64), Expect = 5.3
Identities = 20/77 (25%), Positives = 33/77 (41%)
Frame = +1
Query: 422 AFFDGYINSTTSLNQFVKQYDNALSSRAEKEFEADFNSLDTTIPCVSNSSIEKQLQGEYT 481
A D ++N T Q K+ +++ A K+F + IPC+ N K+L
Sbjct: 88 AMVDDWVNET----QVEKEGSESVTKFAIKKFGLCSRGMSEYIPCLDNVEAIKKLPSTEK 255
Query: 482 HAKFKEVQAEFISKMNC 498
+F+ E K+NC
Sbjct: 256 GERFERHCPEDGKKLNC 306
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,004,061
Number of Sequences: 36976
Number of extensions: 367416
Number of successful extensions: 1818
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1797
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1808
length of query: 688
length of database: 9,014,727
effective HSP length: 103
effective length of query: 585
effective length of database: 5,206,199
effective search space: 3045626415
effective search space used: 3045626415
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146941.15


