

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146941.12 - phase: 0
(146 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
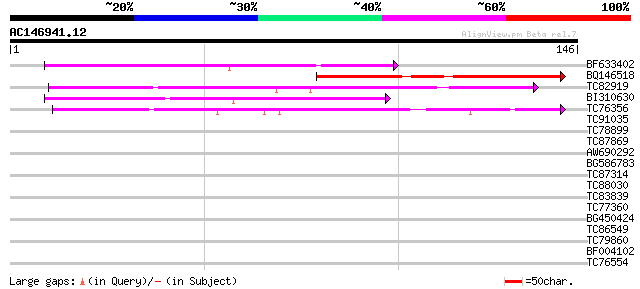
Score E
Sequences producing significant alignments: (bits) Value
BF633402 similar to PIR|C86226|C862 protein T31J12.4 [imported] ... 68 1e-12
BQ146518 weakly similar to PIR|C86226|C862 protein T31J12.4 [imp... 50 4e-07
TC82919 weakly similar to GP|8843783|dbj|BAA97331.1 gb|AAC80581.... 44 2e-05
BI310630 similar to GP|18087548|gb AT5g55600/MDF20_4 {Arabidopsi... 43 5e-05
TC76356 weakly similar to GP|22135896|gb|AAM91530.1 unknown prot... 42 9e-05
TC91035 similar to PIR|C84650|C84650 hypothetical protein At2g25... 36 0.006
TC78899 similar to GP|3900936|emb|CAA10167.1 glucan endo-1 3-bet... 29 0.77
TC87869 similar to GP|23504577|emb|CAD51456. chromosome condensa... 28 1.0
AW690292 weakly similar to GP|22136810|gb| unknown protein {Arab... 27 3.8
BG586783 weakly similar to GP|22135896|gb| unknown protein {Arab... 27 3.8
TC87314 similar to PIR|T00848|T00848 probable serine/threonine-s... 27 3.8
TC88030 similar to PIR|F86416|F86416 probable RNA-binding protei... 27 3.8
TC83839 weakly similar to PIR|T47739|T47739 hypothetical protein... 26 5.0
TC77360 similar to SP|P32110|GTX6_SOYBN Probable glutathione S-t... 26 5.0
BG450424 similar to GP|9294022|dbj| selenium-binding protein-lik... 26 5.0
TC86549 similar to GP|7331143|gb|AAF60293.1| chaperonin 21 precu... 26 6.5
TC79860 homologue to PIR|T06533|T06533 probable gibberellin 20-o... 26 6.5
BF004102 similar to PIR|T47972|T479 hypothetical protein F15G16.... 25 8.5
TC76554 similar to GP|7295688|gb|AAF50994.1| CG11931 gene produc... 25 8.5
>BF633402 similar to PIR|C86226|C862 protein T31J12.4 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 427
Score = 67.8 bits (164), Expect = 1e-12
Identities = 36/92 (39%), Positives = 55/92 (59%), Gaps = 1/92 (1%)
Frame = +1
Query: 10 YKVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDE-SELLMETLFPKDL 68
+K G VE+ S ++GF GS+F IV L + K+++ Y NL++D+ ++ L E+L L
Sbjct: 154 FKPGTLVEISSDDDGFRGSWFTGKIVRRLANNKFMVEYDNLMEDEAGTKHLKESLKLHQL 333
Query: 69 RPLPPRVHNPWRFELNQKVDVFDNDGWWVGEI 100
RP+ P N F+ +VD + NDGWW G I
Sbjct: 334 RPVLPTEANRV-FKFGDEVDAYHNDGWWEGHI 426
>BQ146518 weakly similar to PIR|C86226|C862 protein T31J12.4 [imported] -
Arabidopsis thaliana, partial (11%)
Length = 432
Score = 49.7 bits (117), Expect = 4e-07
Identities = 23/64 (35%), Positives = 40/64 (61%)
Frame = +3
Query: 80 RFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSVYFDYCHQTIYYPCDQIRVHQELVW 139
+FE+ +VD + NDGWWVG ++ K+L + Y V+F ++ I + Q+R+HQ+ +
Sbjct: 21 QFEVLDEVDAYYNDGWWVGVVS--KVLGDSKYI--VFFRNSNEEIEFQDSQLRLHQDWMN 188
Query: 140 GDWI 143
G W+
Sbjct: 189 GKWV 200
>TC82919 weakly similar to GP|8843783|dbj|BAA97331.1
gb|AAC80581.1~gene_id:MZN1.7~similar to unknown protein
{Arabidopsis thaliana}, partial (9%)
Length = 1227
Score = 43.9 bits (102), Expect = 2e-05
Identities = 36/141 (25%), Positives = 64/141 (44%), Gaps = 15/141 (10%)
Frame = +2
Query: 11 KVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKD--- 67
K+ ++VEV S EEGF+GS+ T++ E K ++Y+N+L D+ +E +
Sbjct: 47 KINERVEVKSFEEGFLGSWHPGTVIDS-EKLKRHVQYENILNDNGLGNFVEIVSVSSVLD 223
Query: 68 -----------LRPLPPRVH-NPWRFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSV 115
+RP+PP V + VDV + WW G + + ME+ +V
Sbjct: 224 GDIGSLSKRGRIRPVPPLVEFEKCDLKYGLCVDVNYQEAWWEGVVNDKCDGMEER---TV 394
Query: 116 YFDYCHQTIYYPCDQIRVHQE 136
+F + ++R+ Q+
Sbjct: 395 FFPDLGDEMKVGIQEMRITQD 457
>BI310630 similar to GP|18087548|gb AT5g55600/MDF20_4 {Arabidopsis thaliana},
partial (19%)
Length = 730
Score = 42.7 bits (99), Expect = 5e-05
Identities = 30/105 (28%), Positives = 45/105 (42%), Gaps = 16/105 (15%)
Frame = +1
Query: 10 YKVGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDES------------- 56
YKV DK+EV S++ G G +F TIV + ++Y ++ +D S
Sbjct: 418 YKVDDKIEVLSQDSGIRGCWFRCTIVQVARK-QLKVQYDDVQDEDGSGNLEEWIPAFKLA 594
Query: 57 ---ELLMETLFPKDLRPLPPRVHNPWRFELNQKVDVFDNDGWWVG 98
+L M +RP PP E+ VD + +DGW G
Sbjct: 595 KPDKLEMRQPGRSTIRPAPPLEEQELIVEVGTAVDAWWSDGWGEG 729
>TC76356 weakly similar to GP|22135896|gb|AAM91530.1 unknown protein
{Arabidopsis thaliana}, partial (36%)
Length = 2619
Score = 42.0 bits (97), Expect = 9e-05
Identities = 39/150 (26%), Positives = 67/150 (44%), Gaps = 18/150 (12%)
Frame = +3
Query: 12 VGDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLK-DDESELLMETLF------ 64
VG VEV S++ G G +F A+++ K ++Y ++ +DE+ L E +
Sbjct: 1179 VGSNVEVLSQDSGIRGCWFRASVIK-RHKDKVKVQYHDIQDAEDEANNLEEWILASRPVV 1355
Query: 65 PKDL----------RPLPPRVHNPWRFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYS 114
P DL RPL + + ++ VD + +DGWW G + + E Y
Sbjct: 1356 PDDLGLRVEERTKIRPLLEKRGISFVGDVGYIVDAWWHDGWWEGIVVQK----ESDDKYH 1523
Query: 115 VYF-DYCHQTIYYPCDQIRVHQELVWGDWI 143
VYF +I+ PC+ +R ++ W+
Sbjct: 1524 VYFPGEKKMSIFGPCN-LRHSRDWTGNGWV 1610
>TC91035 similar to PIR|C84650|C84650 hypothetical protein At2g25590
[imported] - Arabidopsis thaliana, partial (7%)
Length = 709
Score = 35.8 bits (81), Expect = 0.006
Identities = 34/138 (24%), Positives = 58/138 (41%), Gaps = 7/138 (5%)
Frame = +2
Query: 13 GDKVEVCSKEEGFVGSYFEATIVSCLESGKYVIRYKNLLKDDESELLMETLFPKDLRPLP 72
G KVE+ +G + A I+S Y + Y N ++ L + K +RP P
Sbjct: 191 GSKVEILVNTQGHGVEWHCARIISG-NGHSYNVEYDN--PSVTAKPLSNRVPRKFIRPCP 361
Query: 73 PRVHNPWRFELNQKVDVFDNDGWWVGEIASEKILMEKSYYYSVYFDYCHQTIYYPCDQIR 132
P + N +E N V+V+D W + ++ M + +Y ++ C +++
Sbjct: 362 PAIENIGSWECNDTVEVWDAGCWKEATVLTD---MTEEFYL--------VRVHGSCMELK 508
Query: 133 VHQELV-----W--GDWI 143
VH+ L W G WI
Sbjct: 509 VHKILTRICQSWQNGQWI 562
>TC78899 similar to GP|3900936|emb|CAA10167.1 glucan endo-1
3-beta-d-glucosidase {Cicer arietinum}, partial (94%)
Length = 1296
Score = 28.9 bits (63), Expect = 0.77
Identities = 11/32 (34%), Positives = 17/32 (52%)
Frame = +2
Query: 111 YYYSVYFDYCHQTIYYPCDQIRVHQELVWGDW 142
Y++S + H T Y C +H+ L+W DW
Sbjct: 68 YHFSACWYTVHWTTIYSCG---IHRSLLWNDW 154
>TC87869 similar to GP|23504577|emb|CAD51456. chromosome condensation
protein putative {Plasmodium falciparum 3D7}, partial
(0%)
Length = 1154
Score = 28.5 bits (62), Expect = 1.0
Identities = 11/27 (40%), Positives = 13/27 (47%)
Frame = +1
Query: 70 PLPPRVHNPWRFELNQKVDVFDNDGWW 96
P PP FE+ VD + DGWW
Sbjct: 1 PRPPESVKGCTFEIGAAVDAWCGDGWW 81
>AW690292 weakly similar to GP|22136810|gb| unknown protein {Arabidopsis
thaliana}, partial (5%)
Length = 679
Score = 26.6 bits (57), Expect = 3.8
Identities = 17/47 (36%), Positives = 23/47 (48%)
Frame = +2
Query: 47 YKNLLKDDESELLMETLFPKDLRPLPPRVHNPWRFELNQKVDVFDND 93
++ +L DDE ELL + DLR LP NQ D+ +ND
Sbjct: 464 FETMLPDDEDELLAGIMDDFDLRRLP-----------NQLEDLDEND 571
>BG586783 weakly similar to GP|22135896|gb| unknown protein {Arabidopsis
thaliana}, partial (12%)
Length = 803
Score = 26.6 bits (57), Expect = 3.8
Identities = 12/38 (31%), Positives = 21/38 (54%)
Frame = +3
Query: 68 LRPLPPRVHNPWRFELNQKVDVFDNDGWWVGEIASEKI 105
+RPL + + ++ VD + +DGWW G S++I
Sbjct: 429 IRPLLEKRGISFVGDVGYIVDAWWHDGWWEGHCCSKRI 542
>TC87314 similar to PIR|T00848|T00848 probable serine/threonine-specific
protein kinase T20F6.6 (EC 2.7.1.-) - Arabidopsis
thaliana, partial (80%)
Length = 1575
Score = 26.6 bits (57), Expect = 3.8
Identities = 11/16 (68%), Positives = 13/16 (80%)
Frame = -3
Query: 121 HQTIYYPCDQIRVHQE 136
HQ+I Y DQIR+HQE
Sbjct: 496 HQSIPYKHDQIRLHQE 449
>TC88030 similar to PIR|F86416|F86416 probable RNA-binding protein MEI2
36123-32976 [imported] - Arabidopsis thaliana, partial
(23%)
Length = 1549
Score = 26.6 bits (57), Expect = 3.8
Identities = 17/47 (36%), Positives = 23/47 (48%)
Frame = +1
Query: 47 YKNLLKDDESELLMETLFPKDLRPLPPRVHNPWRFELNQKVDVFDND 93
++ +L DDE ELL + DLR LP NQ D+ +ND
Sbjct: 610 FETMLPDDEDELLAGIMDDFDLRRLP-----------NQLEDLDEND 717
>TC83839 weakly similar to PIR|T47739|T47739 hypothetical protein F18O21.180
- Arabidopsis thaliana, partial (23%)
Length = 551
Score = 26.2 bits (56), Expect = 5.0
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +1
Query: 6 KRVDYKVGDKVEVCSKEEGFV 26
K VDY K++V KEEGFV
Sbjct: 190 KMVDYDATPKIKVEPKEEGFV 252
>TC77360 similar to SP|P32110|GTX6_SOYBN Probable glutathione S-transferase
(EC 2.5.1.18) (Heat shock protein 26A) (G2-4). [Soybean],
complete
Length = 1140
Score = 26.2 bits (56), Expect = 5.0
Identities = 10/22 (45%), Positives = 14/22 (63%), Gaps = 1/22 (4%)
Frame = -1
Query: 122 QTIYYP-CDQIRVHQELVWGDW 142
+ + YP CD + HQ+ VW DW
Sbjct: 1098 RVLTYPICDYLT*HQQRVWVDW 1033
>BG450424 similar to GP|9294022|dbj| selenium-binding protein-like
{Arabidopsis thaliana}, partial (4%)
Length = 529
Score = 26.2 bits (56), Expect = 5.0
Identities = 13/42 (30%), Positives = 25/42 (58%), Gaps = 6/42 (14%)
Frame = +2
Query: 79 WRFELNQKVDVFDND----GW--WVGEIASEKILMEKSYYYS 114
W FE+ +K+ FD++ W + E+A+EK+L ++ +S
Sbjct: 218 WEFEIGEKIIRFDSECML*SWRNQLAELAAEKVLHLDNHIHS 343
>TC86549 similar to GP|7331143|gb|AAF60293.1| chaperonin 21 precursor
{Lycopersicon esculentum}, partial (92%)
Length = 1134
Score = 25.8 bits (55), Expect = 6.5
Identities = 13/26 (50%), Positives = 18/26 (69%)
Frame = +1
Query: 50 LLKDDESELLMETLFPKDLRPLPPRV 75
+LKDD+ ++ET KDL+PL RV
Sbjct: 502 ILKDDDIVGILETDDIKDLKPLNDRV 579
>TC79860 homologue to PIR|T06533|T06533 probable gibberellin 20-oxidase -
garden pea, partial (93%)
Length = 1454
Score = 25.8 bits (55), Expect = 6.5
Identities = 7/20 (35%), Positives = 10/20 (50%)
Frame = +3
Query: 120 CHQTIYYPCDQIRVHQELVW 139
CH T + PC + E +W
Sbjct: 81 CHDTSFLPCQVTNIPSEFIW 140
>BF004102 similar to PIR|T47972|T479 hypothetical protein F15G16.190 -
Arabidopsis thaliana, partial (9%)
Length = 549
Score = 25.4 bits (54), Expect = 8.5
Identities = 11/24 (45%), Positives = 14/24 (57%)
Frame = -2
Query: 7 RVDYKVGDKVEVCSKEEGFVGSYF 30
+ Y D+V + SKE G VGS F
Sbjct: 404 KFSYLFSDRVPLFSKEHGMVGSQF 333
>TC76554 similar to GP|7295688|gb|AAF50994.1| CG11931 gene product
{Drosophila melanogaster}, partial (42%)
Length = 861
Score = 25.4 bits (54), Expect = 8.5
Identities = 8/16 (50%), Positives = 12/16 (75%)
Frame = +2
Query: 111 YYYSVYFDYCHQTIYY 126
YYYS Y+ YC++ +Y
Sbjct: 533 YYYSYYYYYCYRYHHY 580
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.141 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,769,199
Number of Sequences: 36976
Number of extensions: 82394
Number of successful extensions: 493
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 469
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 486
length of query: 146
length of database: 9,014,727
effective HSP length: 87
effective length of query: 59
effective length of database: 5,797,815
effective search space: 342071085
effective search space used: 342071085
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC146941.12


