

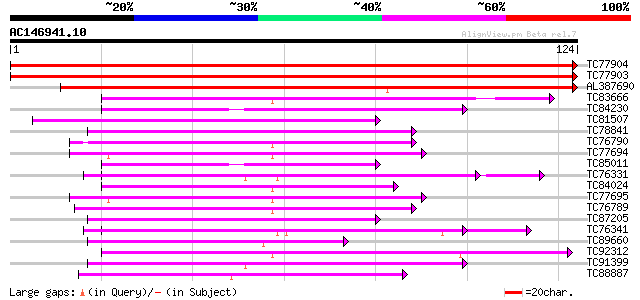
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146941.10 - phase: 0
(124 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77904 homologue to GP|21617984|gb|AAM67034.1 unknown {Arabidop... 249 1e-67
TC77903 homologue to GP|21617984|gb|AAM67034.1 unknown {Arabidop... 249 1e-67
AL387690 homologue to GP|21617984|gb unknown {Arabidopsis thalia... 209 2e-55
TC83666 similar to GP|15042832|gb|AAK82455.1 putative RNA bindin... 52 5e-08
TC84230 51 1e-07
TC81507 similar to GP|22831146|dbj|BAC16007. putative pre-mRNA s... 51 1e-07
TC78841 similar to GP|5815235|gb|AAD52609.1| splicing factor SR1... 49 4e-07
TC76790 similar to PIR|T06817|T06817 RNA-binding protein - garde... 49 7e-07
TC77694 homologue to PIR|T09704|T09704 probable arginine/serine-... 47 2e-06
TC85011 47 2e-06
TC76331 similar to GP|7673355|gb|AAF66823.1| poly(A)-binding pro... 47 2e-06
TC84024 similar to PIR|S77714|S77714 RNA-binding protein precurs... 46 3e-06
TC77695 similar to PIR|T09704|T09704 probable arginine/serine-ri... 46 4e-06
TC76789 similar to GP|15294254|gb|AAK95304.1 AT4g24770/F22K18_30... 45 6e-06
TC87205 similar to GP|12324445|gb|AAG52185.1 putative splicing f... 44 2e-05
TC76341 similar to GP|7528270|gb|AAF63202.1| poly(A)-binding pro... 43 3e-05
TC89660 similar to GP|11034579|dbj|BAB17103. hypothetical protei... 43 3e-05
TC92312 weakly similar to GP|4567275|gb|AAD23688.1| expressed pr... 43 4e-05
TC91399 weakly similar to PIR|S53494|S53494 RNA-binding protein ... 42 5e-05
TC88887 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproli... 42 5e-05
>TC77904 homologue to GP|21617984|gb|AAM67034.1 unknown {Arabidopsis
thaliana}, complete
Length = 695
Score = 249 bits (637), Expect = 1e-67
Identities = 124/124 (100%), Positives = 124/124 (100%)
Frame = +3
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA
Sbjct: 57 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 236
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 120
FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS
Sbjct: 237 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 416
Query: 121 TKDK 124
TKDK
Sbjct: 417 TKDK 428
>TC77903 homologue to GP|21617984|gb|AAM67034.1 unknown {Arabidopsis
thaliana}, complete
Length = 802
Score = 249 bits (637), Expect = 1e-67
Identities = 124/124 (100%), Positives = 124/124 (100%)
Frame = +2
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA
Sbjct: 56 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 235
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 120
FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS
Sbjct: 236 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 415
Query: 121 TKDK 124
TKDK
Sbjct: 416 TKDK 427
>AL387690 homologue to GP|21617984|gb unknown {Arabidopsis thaliana}, partial
(86%)
Length = 389
Score = 209 bits (533), Expect = 2e-55
Identities = 107/116 (92%), Positives = 107/116 (92%), Gaps = 3/116 (2%)
Frame = +2
Query: 12 RLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAK 71
RLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAK
Sbjct: 2 RLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAK 181
Query: 72 TAVDHLSGFN---VANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVSTKDK 124
TAVDHLS RYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVSTKDK
Sbjct: 182 TAVDHLSXVQSGXXLTRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVSTKDK 349
>TC83666 similar to GP|15042832|gb|AAK82455.1 putative RNA binding protein
{Oryza sativa}, partial (52%)
Length = 896
Score = 52.4 bits (124), Expect = 5e-08
Identities = 31/102 (30%), Positives = 55/102 (53%), Gaps = 3/102 (2%)
Frame = +1
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDAKTAVDHL 77
++V +PF+ T ++ +F +YG + I + +K T +G AF+ YED AVD+L
Sbjct: 274 VFVGGIPFDFTEGDVIAVFAQYGEVVDINLVRDKGTGKSKGFAFIAYEDQRSTNLAVDNL 453
Query: 78 SGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGV 119
+G V+ R + V + + K ++ D E+E + +E GV
Sbjct: 454 NGAQVSGRIIRVDHVDKYKKMEEED----EEEAKQKREARGV 567
>TC84230
Length = 691
Score = 51.2 bits (121), Expect = 1e-07
Identities = 29/80 (36%), Positives = 45/80 (56%)
Frame = +2
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAKTAVDHLSGF 80
LYV NL ++T ++ D+F +YGA+ + T+ R AFV ++ I DAK A + L GF
Sbjct: 224 LYVANLSPDVTDSDLMDLFVQYGALDSV---TSYSARNYAFVFFKRIDDAKAAKNALQGF 394
Query: 81 NVANRYLIVLYYQQAKMSKK 100
N L + + + AK K+
Sbjct: 395 NFRGNSLRIEFARPAKTCKQ 454
>TC81507 similar to GP|22831146|dbj|BAC16007. putative pre-mRNA splicing
factor SF2 (SR1 protein) {Oryza sativa (japonica
cultivar-group)}, partial (57%)
Length = 1287
Score = 50.8 bits (120), Expect = 1e-07
Identities = 27/76 (35%), Positives = 41/76 (53%)
Frame = +2
Query: 6 LRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYE 65
L+ R+ +R +YV NLP +I E+ D+F KYG I I + G AFV +E
Sbjct: 71 LKSKKRRMSRHSSRTVYVGNLPGDIREREVEDLFMKYGHITHIDLKVPPRPPGYAFVEFE 250
Query: 66 DIYDAKTAVDHLSGFN 81
D+ DA+ A+ G++
Sbjct: 251 DVQDAEDAIRGRDGYD 298
>TC78841 similar to GP|5815235|gb|AAD52609.1| splicing factor SR1
{Arabidopsis thaliana}, partial (74%)
Length = 1380
Score = 49.3 bits (116), Expect = 4e-07
Identities = 28/72 (38%), Positives = 40/72 (54%)
Frame = +3
Query: 18 NRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAKTAVDHL 77
+R LYV NLP +I E+ D+F KYG I I + G AFV +ED DA+ A+ +
Sbjct: 57 SRTLYVGNLPGDIRLREVEDLFYKYGPIVDIDLKIPPKPPGYAFVEFEDARDAQDAIYYR 236
Query: 78 SGFNVANRYLIV 89
G++ L+V
Sbjct: 237 DGYDFDGYRLLV 272
>TC76790 similar to PIR|T06817|T06817 RNA-binding protein - garden pea,
partial (79%)
Length = 1141
Score = 48.5 bits (114), Expect = 7e-07
Identities = 26/79 (32%), Positives = 46/79 (57%), Gaps = 3/79 (3%)
Frame = +1
Query: 14 PPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDA 70
PPE + L+V N PF++ SE++ +FG+ G + + N+ T RG FV + +A
Sbjct: 349 PPE-DAKLFVGNFPFDVDSEKLAMLFGQAGTVEIAEVIYNRQTDLSRGFGFVTMSTVEEA 525
Query: 71 KTAVDHLSGFNVANRYLIV 89
++AV+ +G++ R L+V
Sbjct: 526 ESAVEKFNGYDYNGRSLVV 582
>TC77694 homologue to PIR|T09704|T09704 probable arginine/serine-rich
splicing factor - alfalfa, partial (95%)
Length = 1494
Score = 47.0 bits (110), Expect = 2e-06
Identities = 29/83 (34%), Positives = 46/83 (54%), Gaps = 5/83 (6%)
Frame = +3
Query: 14 PPEVNRV--LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIY 68
PP+++ L V N+ F T+++++ +F KYG + I I ++ T RG AFV Y+
Sbjct: 63 PPDISDTYSLLVLNITFRTTADDLFPLFDKYGKVVDIFIPKDRRTGESRGFAFVRYKYAD 242
Query: 69 DAKTAVDHLSGFNVANRYLIVLY 91
+A AVD L G V R + V +
Sbjct: 243 EASKAVDRLDGRMVDGREITVQF 311
>TC85011
Length = 498
Score = 47.0 bits (110), Expect = 2e-06
Identities = 25/61 (40%), Positives = 37/61 (59%)
Frame = +2
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAKTAVDHLSGF 80
LYV NL ++T ++ D+F +YGA+ + T+ R AFV ++ I DAK A + L GF
Sbjct: 161 LYVANLSPDVTDSDLMDLFVQYGALDSV---TSYSARNYAFVFFKRIDDAKAAKNALQGF 331
Query: 81 N 81
N
Sbjct: 332 N 334
>TC76331 similar to GP|7673355|gb|AAF66823.1| poly(A)-binding protein
{Nicotiana tabacum}, partial (97%)
Length = 2581
Score = 46.6 bits (109), Expect = 2e-06
Identities = 27/90 (30%), Positives = 47/90 (52%), Gaps = 3/90 (3%)
Frame = +3
Query: 17 VNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTR---GTAFVVYEDIYDAKTA 73
V LYV +L N+T ++YD+F + G + +R+ + TR G +V Y + DA A
Sbjct: 438 VTTSLYVGDLDMNVTDSQLYDLFNQLGQVVSVRVCRDLTTRRSLGYGYVNYSNPQDAARA 617
Query: 74 VDHLSGFNVANRYLIVLYYQQAKMSKKFDQ 103
+D L+ + NR + ++Y + +K Q
Sbjct: 618 LDVLNFTPLNNRPIRIMYSHRDPSIRKSGQ 707
Score = 46.2 bits (108), Expect = 3e-06
Identities = 30/99 (30%), Positives = 51/99 (51%), Gaps = 2/99 (2%)
Frame = +3
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRI--GTNKDTRGTAFVVYEDIYDAKTAVDHLS 78
LYV+NL +I E++ ++F YG I ++ N +RG+ FV + +A A+ ++
Sbjct: 1296 LYVKNLDDSIADEKLKELFSSYGTITSCKVMRDPNGVSRGSGFVAFSTPEEASRALLEMN 1475
Query: 79 GFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKY 117
G VA++ L V Q +KED AR+Q ++
Sbjct: 1476 GKMVASKPLYVTLAQ-----------RKEDRRARLQAQF 1559
>TC84024 similar to PIR|S77714|S77714 RNA-binding protein precursor 33K -
wood tobacco, partial (55%)
Length = 867
Score = 46.2 bits (108), Expect = 3e-06
Identities = 26/68 (38%), Positives = 38/68 (55%), Gaps = 3/68 (4%)
Frame = +3
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDAKTAVDHL 77
LYV NLPF++TS ++ DIF + G + + I ++ T RG AFV + +AK A+
Sbjct: 339 LYVGNLPFSMTSSQLADIFAEAGTVVSVEIVYDRLTDRSRGFAFVTMKSAEEAKEAIRMF 518
Query: 78 SGFNVANR 85
G V R
Sbjct: 519 DGSQVGGR 542
>TC77695 similar to PIR|T09704|T09704 probable arginine/serine-rich splicing
factor - alfalfa, partial (83%)
Length = 1657
Score = 45.8 bits (107), Expect = 4e-06
Identities = 29/83 (34%), Positives = 45/83 (53%), Gaps = 5/83 (6%)
Frame = +1
Query: 14 PPEVNRV--LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIY 68
PP++ L V N+ F T+++++ +F KYG + I I ++ T RG AFV Y+
Sbjct: 91 PPDIADTYSLLVLNITFRTTADDLFPLFDKYGKVVDIFIPRDRRTGESRGFAFVRYKYAD 270
Query: 69 DAKTAVDHLSGFNVANRYLIVLY 91
+A AVD L G V R + V +
Sbjct: 271 EASKAVDRLDGRMVDGREITVQF 339
>TC76789 similar to GP|15294254|gb|AAK95304.1 AT4g24770/F22K18_30
{Arabidopsis thaliana}, partial (62%)
Length = 1294
Score = 45.4 bits (106), Expect = 6e-06
Identities = 25/78 (32%), Positives = 41/78 (52%), Gaps = 3/78 (3%)
Frame = +3
Query: 15 PEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDAK 71
P + ++V NLPF++ SE++ +F + G + + N+DT RG FV + +
Sbjct: 441 PSEDLKIFVGNLPFDVDSEKLAQLFEQSGTVEIAEVIYNRDTDRSRGFGFVTMSTSEEVE 620
Query: 72 TAVDHLSGFNVANRYLIV 89
AV+ SGF + R L V
Sbjct: 621 RAVNKFSGFELDGRLLTV 674
>TC87205 similar to GP|12324445|gb|AAG52185.1 putative splicing factor;
53460-55514 {Arabidopsis thaliana}, partial (81%)
Length = 1197
Score = 43.5 bits (101), Expect = 2e-05
Identities = 22/64 (34%), Positives = 35/64 (54%)
Frame = +2
Query: 18 NRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAKTAVDHL 77
+R +YV NLP +I E+ D+F KYG I +I + FV +++ DA+ A+
Sbjct: 116 SRTIYVGNLPADIRESEIEDLFYKYGRIMEIELKVPPRPPCYCFVEFDNARDAEDAIRGR 295
Query: 78 SGFN 81
G+N
Sbjct: 296 DGYN 307
>TC76341 similar to GP|7528270|gb|AAF63202.1| poly(A)-binding protein
{Cucumis sativus}, partial (90%)
Length = 2501
Score = 43.1 bits (100), Expect = 3e-05
Identities = 25/87 (28%), Positives = 46/87 (52%), Gaps = 3/87 (3%)
Frame = +3
Query: 17 VNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTR---GTAFVVYEDIYDAKTA 73
V LYV +L N+ ++YD+F + G + +R+ + TR G +V + + DA A
Sbjct: 288 VTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARA 467
Query: 74 VDHLSGFNVANRYLIVLYYQQAKMSKK 100
+D L+ + N+ + V+Y + S+K
Sbjct: 468 LDVLNFTPMNNKSIRVMYSHRDPSSRK 548
Score = 41.6 bits (96), Expect = 8e-05
Identities = 29/101 (28%), Positives = 51/101 (49%), Gaps = 7/101 (6%)
Frame = +3
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGT--AFVVYEDIYDAKTAVDHLS 78
+YV+NL + T +++ + FG YG I + + D R FV +E+ DA AV+ L+
Sbjct: 837 VYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALN 1016
Query: 79 GFNVANRYLIVLYYQ-----QAKMSKKFDQKKKEDEIARMQ 114
G V ++ V Q + ++ +F+Q KE + + Q
Sbjct: 1017GKKVDDKEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQ 1139
>TC89660 similar to GP|11034579|dbj|BAB17103. hypothetical protein~similar
to Arabidopsis thaliana chromosome 3 K15M2.15, partial
(16%)
Length = 1035
Score = 43.1 bits (100), Expect = 3e-05
Identities = 23/60 (38%), Positives = 36/60 (59%), Gaps = 3/60 (5%)
Frame = +3
Query: 18 NRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNK---DTRGTAFVVYEDIYDAKTAV 74
+R ++V N+PF +++E + F KYG I + +G +K TRG AF VY+ AK +V
Sbjct: 24 SRKVFVGNVPFEVSAESLLSEFSKYGEIEEGPLGFDKASGKTRGFAFFVYKTEEGAKNSV 203
>TC92312 weakly similar to GP|4567275|gb|AAD23688.1| expressed protein
{Arabidopsis thaliana}, partial (29%)
Length = 1319
Score = 42.7 bits (99), Expect = 4e-05
Identities = 31/108 (28%), Positives = 51/108 (46%), Gaps = 5/108 (4%)
Frame = +1
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDAKTAVDHL 77
L VRNLPF E+ D F G + ++ I DT +G AFV + DA+ A+ L
Sbjct: 280 LIVRNLPFKAKENEIRDAFSSAGTVWEVFIPQKSDTGLSKGFAFVKFTCKQDAENAIRKL 459
Query: 78 SGFNVANRYLIVLYYQQAKM--SKKFDQKKKEDEIARMQEKYGVSTKD 123
+G +R + V + K+ S D E+ ++ ++ G +T +
Sbjct: 460 NGSKFGSRLIAVDWAVPKKIFSSDTNDAPASEEGQQKVTDEDGSTTTE 603
>TC91399 weakly similar to PIR|S53494|S53494 RNA-binding protein cp33
precursor - Arabidopsis thaliana, partial (39%)
Length = 775
Score = 42.4 bits (98), Expect = 5e-05
Identities = 25/86 (29%), Positives = 45/86 (52%), Gaps = 3/86 (3%)
Frame = +2
Query: 18 NRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRI---GTNKDTRGTAFVVYEDIYDAKTAV 74
+R L V NLPF+++S ++ +FG+ G + + I +RG AFV ++ DA+ A+
Sbjct: 245 SRRLLVGNLPFSLSSSQLAQLFGEAGNVVSVEILYDDITNRSRGFAFVTMGNVEDAEEAI 424
Query: 75 DHLSGFNVANRYLIVLYYQQAKMSKK 100
G V R + V + + + K+
Sbjct: 425 RMFDGTTVGGRAIKVNFPEVPIVGKR 502
>TC88887 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich
glycoprotein DZ-HRGP {Volvox carteri f. nagariensis},
partial (18%)
Length = 1681
Score = 42.4 bits (98), Expect = 5e-05
Identities = 22/74 (29%), Positives = 40/74 (53%), Gaps = 2/74 (2%)
Frame = -1
Query: 16 EVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQ--IRIGTNKDTRGTAFVVYEDIYDAKTA 73
E +YVRNLP++ ++E++ ++F G + Q I+ + +RGT V ++ A+TA
Sbjct: 496 ERGETIYVRNLPWSTSNEDLVELFTTIGKVEQAEIQYEPSGRSRGTGVVRFDSAETAETA 317
Query: 74 VDHLSGFNVANRYL 87
+ G+ R L
Sbjct: 316 IAKFQGYQYGGRPL 275
Score = 37.4 bits (85), Expect = 0.001
Identities = 18/73 (24%), Positives = 40/73 (54%), Gaps = 5/73 (6%)
Frame = -2
Query: 18 NRVLYVRNLPFNITSEEMYDIF---GKYGAI--RQIRIGTNKDTRGTAFVVYEDIYDAKT 72
+R ++V NLP+ + +++ D+F + G + + +G + +G+ VV+E+ DA+
Sbjct: 981 SRQIFVANLPYTVGWQDLKDLFRQAARNGVVIRADVHLGPDGRPKGSGIVVFENPEDARN 802
Query: 73 AVDHLSGFNVANR 85
A+ G++ R
Sbjct: 801 AIQQFHGYDWQGR 763
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,627,352
Number of Sequences: 36976
Number of extensions: 24242
Number of successful extensions: 251
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 240
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 245
length of query: 124
length of database: 9,014,727
effective HSP length: 84
effective length of query: 40
effective length of database: 5,908,743
effective search space: 236349720
effective search space used: 236349720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC146941.10


