

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146940.1 - phase: 0
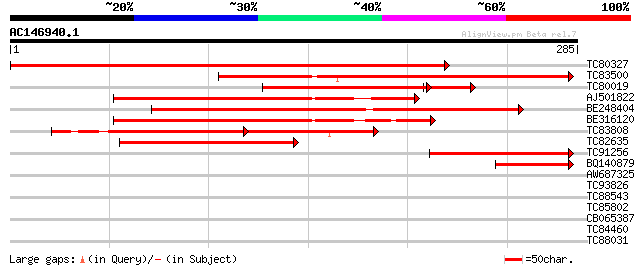
(285 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80327 similar to GP|3540182|gb|AAC34332.1| Unknown protein {Ar... 453 e-128
TC83500 similar to PIR|T04231|T04231 hypothetical protein F14M19... 191 4e-49
TC80019 similar to PIR|G86446|G86446 unknown protein [imported] ... 158 2e-39
AJ501822 similar to GP|20258913|gb unknown protein {Arabidopsis ... 155 2e-38
BE248404 similar to GP|20260538|gb unknown protein {Arabidopsis ... 151 3e-37
BE316120 similar to GP|20258913|gb unknown protein {Arabidopsis ... 139 2e-33
TC83808 similar to PIR|T04231|T04231 hypothetical protein F14M19... 99 3e-33
TC82635 similar to GP|20260538|gb|AAM13167.1 unknown protein {Ar... 92 3e-19
TC91256 similar to GP|20258913|gb|AAM14150.1 unknown protein {Ar... 72 3e-13
BQ140879 similar to GP|20260538|gb unknown protein {Arabidopsis ... 48 4e-06
AW687325 similar to GP|21689821|gb unknown protein {Arabidopsis ... 33 0.096
TC93826 similar to PIR|H86366|H86366 protein F26F24.12 [imported... 32 0.36
TC88543 similar to GP|20466782|gb|AAM20708.1 unknown protein {Ar... 31 0.62
TC85802 weakly similar to PIR|T47837|T47837 beta-glucosidase-lik... 29 1.8
CB065387 weakly similar to GP|20257161|gb| 2-dehydro-3-deoxygala... 29 1.8
TC84460 similar to PIR|T47960|T47960 hypothetical protein F15G16... 28 4.0
TC88031 similar to PIR|E84706|E84706 hypothetical protein At2g30... 27 6.9
>TC80327 similar to GP|3540182|gb|AAC34332.1| Unknown protein {Arabidopsis
thaliana}, partial (34%)
Length = 822
Score = 453 bits (1166), Expect = e-128
Identities = 220/221 (99%), Positives = 220/221 (99%)
Frame = +1
Query: 1 MESQQIPKVQHSVVDVQINNKNIKKLKFPKFGCFRIQHDATGDGFDIEVVDASGHRSNPT 60
MESQQIPKVQHSVVDVQINNKNIKKLKFPKFGCFRIQHDATGDGFDIEVVDASGHRSNPT
Sbjct: 154 MESQQIPKVQHSVVDVQINNKNIKKLKFPKFGCFRIQHDATGDGFDIEVVDASGHRSNPT 333
Query: 61 HLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLAEEVI 120
HLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLAEEVI
Sbjct: 334 HLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLAEEVI 513
Query: 121 SVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQISNQECHVRK 180
SVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQISNQECHVRK
Sbjct: 514 SVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQISNQECHVRK 693
Query: 181 YEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEKT 221
YEGKIAGLEPINFITSATPHLGCRGHKQVPL CGFHSLEKT
Sbjct: 694 YEGKIAGLEPINFITSATPHLGCRGHKQVPLXCGFHSLEKT 816
>TC83500 similar to PIR|T04231|T04231 hypothetical protein F14M19.50 -
Arabidopsis thaliana, partial (29%)
Length = 994
Score = 191 bits (484), Expect = 4e-49
Identities = 100/184 (54%), Positives = 131/184 (70%), Gaps = 6/184 (3%)
Frame = +2
Query: 106 DGVDVTGNRLAEEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQG-- 163
DGVD EEV+S+++ P ++KISF+AHSLGGL+ARYAIA+L+ D SK L G
Sbjct: 2 DGVDTWVRG*PEEVLSIVRCWPGLQKISFVAHSLGGLVARYAIARLF--DYSKTLEAGVT 175
Query: 164 --NVHCEGQIS-NQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEK 220
N C+ + + C + YE +IAGLEP+NFIT ATPHLG RGH+Q+P LCG LE+
Sbjct: 176 CRNCDCKEEAECTKNCTEQHYEARIAGLEPMNFITFATPHLGSRGHRQLPFLCGIPFLER 355
Query: 221 TASRLSRFL-GKTGKHLFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRY 279
AS+ + + G+TGKHLFL D + KPPLLL+M+ DS+D+KFMSAL FKRRVAYAN +
Sbjct: 356 RASQTAHLIVGRTGKHLFLMDNDDGKPPLLLRMIEDSDDLKFMSALCVFKRRVAYANANF 535
Query: 280 DRIL 283
D ++
Sbjct: 536 DHMV 547
>TC80019 similar to PIR|G86446|G86446 unknown protein [imported] -
Arabidopsis thaliana, partial (35%)
Length = 1518
Score = 158 bits (400), Expect = 2e-39
Identities = 77/85 (90%), Positives = 80/85 (93%)
Frame = +2
Query: 128 SVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQISNQECHVRKYEGKIAG 187
SV+KISFIAHSLGGLIARYAIAKLYERDISKELSQGNVH E QISNQECH+RKYEGKIAG
Sbjct: 674 SVQKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHSESQISNQECHIRKYEGKIAG 853
Query: 188 LEPINFITSATPHLGCRGHKQVPLL 212
LEPINFITS PHLGCRGHKQ+ LL
Sbjct: 854 LEPINFITSTMPHLGCRGHKQLILL 928
Score = 42.7 bits (99), Expect = 2e-04
Identities = 20/26 (76%), Positives = 21/26 (79%)
Frame = +3
Query: 209 VPLLCGFHSLEKTASRLSRFLGKTGK 234
VPL+CGF SLEKT SRLSRF GK K
Sbjct: 1167 VPLVCGFDSLEKTTSRLSRFFGKNIK 1244
>AJ501822 similar to GP|20258913|gb unknown protein {Arabidopsis thaliana},
partial (41%)
Length = 597
Score = 155 bits (391), Expect = 2e-38
Identities = 81/154 (52%), Positives = 107/154 (68%)
Frame = +3
Query: 53 SGHRSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTG 112
S S+ HL++MVNG++GS+ +WK+A++QF+K P V VHCSE N S T DGVDV G
Sbjct: 162 SSDSSSADHLVVMVNGILGSSTDWKFASEQFVKELPDKVFVHCSERNVSKHTLDGVDVMG 341
Query: 113 NRLAEEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQIS 172
RLAEEVI VI+R P++RK+SFI+HS+GGL+ARYAI KLY R E Q + + E ++
Sbjct: 342 ERLAEEVIEVIRRKPNMRKVSFISHSVGGLVARYAIGKLY-RPPGNEPIQDSGNKESKVD 518
Query: 173 NQECHVRKYEGKIAGLEPINFITSATPHLGCRGH 206
+ G I GLE +NF+T ATPHLG RG+
Sbjct: 519 S--------IGTICGLEAMNFVTVATPHLGSRGN 596
>BE248404 similar to GP|20260538|gb unknown protein {Arabidopsis thaliana},
partial (32%)
Length = 571
Score = 151 bits (381), Expect = 3e-37
Identities = 87/190 (45%), Positives = 116/190 (60%), Gaps = 3/190 (1%)
Frame = +1
Query: 72 SAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLAEEVISVIKRHPSVRK 131
S +W YA ++ ++H S N+ T TF G+D G RLA+EV+ V+K++ S+++
Sbjct: 10 SPGDWSYAEEELKMNLGKSFLIHASSSNAYTKTFTGIDEAGKRLADEVMQVVKKNQSLKR 189
Query: 132 ISFIAHSLGGLIARYAIAKLYERD-ISKELSQGNVHCEGQISNQECHVRKYEGKIAGLEP 190
ISF+AHSLGGL ARYAIA LY D + V+CE + S + R G IAGLEP
Sbjct: 190 ISFLAHSLGGLFARYAIAVLYSPDTYNSGQPDDPVNCEMENSQKTDFSR---GMIAGLEP 360
Query: 191 INFITSATPHLGCRGHKQVPLLCGFHSLEKTASRLS-RFLGKTGKHLFLTDGKNEKPPLL 249
+NFIT ATPHLG RG Q+P L G LEK + ++ F+G+TG LFLTD K KP L
Sbjct: 361 MNFITLATPHLGVRGKNQLPFLFGVPILEKLVAPVAPLFIGRTGSQLFLTDDKPNKPSLS 540
Query: 250 L-QMVRDSED 258
+ + D ED
Sbjct: 541 F*EWLSDCED 570
>BE316120 similar to GP|20258913|gb unknown protein {Arabidopsis thaliana},
partial (43%)
Length = 625
Score = 139 bits (349), Expect = 2e-33
Identities = 79/162 (48%), Positives = 105/162 (64%)
Frame = +2
Query: 53 SGHRSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTG 112
S S+ HL++MVNG++GS+ +WK+A++QF+K P V VHCSE N S T DGVDV G
Sbjct: 158 SSDSSSADHLVVMVNGILGSSTDWKFASEQFVKELPDKVFVHCSERNVSKHTLDGVDVMG 337
Query: 113 NRLAEEVISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKELSQGNVHCEGQIS 172
RLAEEVI VI+R P++RK+SFI+HS+GGL+ARYAI KLY R E Q + + E ++
Sbjct: 338 ERLAEEVIEVIRRKPNMRKVSFISHSVGGLVARYAIGKLY-RPPGNEPIQDSGNKESKVD 514
Query: 173 NQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCG 214
+ +Y + G E N T LG G+KQVP L G
Sbjct: 515 S-----NRYNMRPGGNEFRN--CCYTSSLGQGGNKQVPFLFG 619
>TC83808 similar to PIR|T04231|T04231 hypothetical protein F14M19.50 -
Arabidopsis thaliana, partial (24%)
Length = 685
Score = 99.0 bits (245), Expect(2) = 3e-33
Identities = 51/99 (51%), Positives = 69/99 (69%)
Frame = +2
Query: 22 NIKKLKFPKFGCFRIQHDATGDGFDIEVVDASGHRSNPTHLIIMVNGLIGSAHNWKYAAK 81
N K++ P+ +++ + +G+ F +AS + P HL+IMVNG+ GSA +W+YAA+
Sbjct: 176 NSIKMQLPRL---KVEAEISGEDF----FNASTSKRIPRHLVIMVNGITGSASDWRYAAE 334
Query: 82 QFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRLAEEVI 120
QF+KR P VIVH SECNSS LTFDGVD G RLAEEV+
Sbjct: 335 QFVKRLPDKVIVHRSECNSSRLTFDGVDTMGERLAEEVL 451
Score = 60.1 bits (144), Expect(2) = 3e-33
Identities = 31/70 (44%), Positives = 45/70 (64%), Gaps = 3/70 (4%)
Frame = +1
Query: 119 VISVIKRHPSVRKISFIAHSLGGLIARYAIAKLYERDISKE---LSQGNVHCEGQISNQE 175
V+SV++R P V KISF+AHSLGGL+ARYAI +LY+ E S+ + E +++
Sbjct: 454 VLSVVRRWPEVHKISFVAHSLGGLVARYAIGRLYDNSSKLEHVGNSRNHFKEEKTEYSKQ 633
Query: 176 CHVRKYEGKI 185
C + YE K+
Sbjct: 634 CLTQSYEAKL 663
>TC82635 similar to GP|20260538|gb|AAM13167.1 unknown protein {Arabidopsis
thaliana}, partial (21%)
Length = 611
Score = 91.7 bits (226), Expect = 3e-19
Identities = 40/90 (44%), Positives = 64/90 (70%)
Frame = +2
Query: 56 RSNPTHLIIMVNGLIGSAHNWKYAAKQFLKRYPYDVIVHCSECNSSTLTFDGVDVTGNRL 115
R++P HL+++V+G++ S +W YA + KR + +++ S N+ T TF G+D G RL
Sbjct: 341 RNDPDHLLVLVHGILASTADWTYAEAELKKRLGKNFLIYVSSSNAYTKTFTGIDGAGKRL 520
Query: 116 AEEVISVIKRHPSVRKISFIAHSLGGLIAR 145
A+EV+ V+K+ S+++ISF+AHSLGGL AR
Sbjct: 521 ADEVLQVVKKTESLKRISFLAHSLGGLFAR 610
>TC91256 similar to GP|20258913|gb|AAM14150.1 unknown protein {Arabidopsis
thaliana}, partial (48%)
Length = 675
Score = 71.6 bits (174), Expect = 3e-13
Identities = 37/73 (50%), Positives = 50/73 (67%), Gaps = 1/73 (1%)
Frame = +2
Query: 212 LCGFHSLEKTASRLSRFL-GKTGKHLFLTDGKNEKPPLLLQMVRDSEDIKFMSALRSFKR 270
L G + EK AS + ++ +TG+HLFLTD KPPLL +M+ D + FMSALR+FKR
Sbjct: 5 LFGVTAFEKLASVVIHWIFRRTGRHLFLTDDDEGKPPLLKRMIEDYDGYYFMSALRTFKR 184
Query: 271 RVAYANIRYDRIL 283
RV Y+N+ YD I+
Sbjct: 185 RVIYSNVGYDHIV 223
>BQ140879 similar to GP|20260538|gb unknown protein {Arabidopsis thaliana},
partial (36%)
Length = 618
Score = 48.1 bits (113), Expect = 4e-06
Identities = 22/39 (56%), Positives = 29/39 (73%)
Frame = +1
Query: 245 KPPLLLQMVRDSEDIKFMSALRSFKRRVAYANIRYDRIL 283
KP LLL+M D ED KF+SAL +F+ RV YAN+ YD ++
Sbjct: 4 KPSLLLRMASDCEDGKFISALGAFRSRVVYANVSYDHMV 120
>AW687325 similar to GP|21689821|gb unknown protein {Arabidopsis thaliana},
partial (19%)
Length = 505
Score = 33.5 bits (75), Expect = 0.096
Identities = 19/45 (42%), Positives = 26/45 (57%), Gaps = 9/45 (20%)
Frame = +3
Query: 114 RLAEEVISVIKRHPSVR---------KISFIAHSLGGLIARYAIA 149
RLA+EVIS +K+ ++SF+ HS+G LI R AIA
Sbjct: 6 RLAQEVISFVKKKMDKESRCGNLRDIRLSFVGHSIGNLIIRTAIA 140
>TC93826 similar to PIR|H86366|H86366 protein F26F24.12 [imported] -
Arabidopsis thaliana, partial (56%)
Length = 672
Score = 31.6 bits (70), Expect = 0.36
Identities = 19/64 (29%), Positives = 29/64 (44%), Gaps = 3/64 (4%)
Frame = +2
Query: 164 NVHCEGQI---SNQECHVRKYEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFHSLEK 220
N+ EG+I S+Q C K GK++G +++ T + CR V +C S
Sbjct: 29 NLKTEGKILKISSQRCSTMKLYGKLSGTNHCSYMAKITTGIFCRNPYNVTGICNRSSCPL 208
Query: 221 TASR 224
SR
Sbjct: 209 ANSR 220
>TC88543 similar to GP|20466782|gb|AAM20708.1 unknown protein {Arabidopsis
thaliana}, partial (46%)
Length = 2101
Score = 30.8 bits (68), Expect = 0.62
Identities = 20/67 (29%), Positives = 33/67 (48%), Gaps = 2/67 (2%)
Frame = +1
Query: 130 RKISFIAHSLGGLIARYAI--AKLYERDISKELSQGNVHCEGQISNQECHVRKYEGKIAG 187
+K + HSL +I R ++ A+ E + K L+ HCE + V + GKIA
Sbjct: 844 KKQYLLLHSLKEVIVRQSVDKAEFQESSVEKILNLLFNHCESEEEGVRNVVAECLGKIAL 1023
Query: 188 LEPINFI 194
+EP+ +
Sbjct: 1024IEPVKLV 1044
>TC85802 weakly similar to PIR|T47837|T47837 beta-glucosidase-like protein -
Arabidopsis thaliana, partial (43%)
Length = 1119
Score = 29.3 bits (64), Expect = 1.8
Identities = 11/26 (42%), Positives = 17/26 (65%)
Frame = -1
Query: 59 PTHLIIMVNGLIGSAHNWKYAAKQFL 84
PT+L I +N LI + HNW + Q++
Sbjct: 1116 PTNLFIDINLLISALHNWYHQLSQYI 1039
>CB065387 weakly similar to GP|20257161|gb| 2-dehydro-3-deoxygalactonate
kinase {Bradyrhizobium japonicum}, partial (16%)
Length = 633
Score = 29.3 bits (64), Expect = 1.8
Identities = 17/46 (36%), Positives = 24/46 (51%), Gaps = 1/46 (2%)
Frame = -3
Query: 36 IQHDATGDGFDIEVVDASGHRSNPT-HLIIMVNGLIGSAHNWKYAA 80
I A DGF++ DA G + L ++ G+IGSA W+ AA
Sbjct: 553 IAGQACSDGFELAFDDACGDWLDAQPELPVIACGMIGSAQGWREAA 416
>TC84460 similar to PIR|T47960|T47960 hypothetical protein F15G16.70 -
Arabidopsis thaliana, partial (23%)
Length = 843
Score = 28.1 bits (61), Expect = 4.0
Identities = 11/27 (40%), Positives = 17/27 (62%)
Frame = +2
Query: 118 EVISVIKRHPSVRKISFIAHSLGGLIA 144
E++ +KRH K+ F HSLGG ++
Sbjct: 578 EIMDHLKRHGDRAKLQFTGHSLGGSLS 658
>TC88031 similar to PIR|E84706|E84706 hypothetical protein At2g30280
[imported] - Arabidopsis thaliana, partial (11%)
Length = 1443
Score = 27.3 bits (59), Expect = 6.9
Identities = 15/55 (27%), Positives = 23/55 (41%), Gaps = 4/55 (7%)
Frame = -3
Query: 166 HCEGQISNQECHVRK----YEGKIAGLEPINFITSATPHLGCRGHKQVPLLCGFH 216
H Q N C + ++ ++ NF+ + L CRGHK PL+ H
Sbjct: 853 HLSRQFHNLNCQAHQNNLLHQLELVDKNVKNFLQRLSHPLQCRGHKHSPLVWNDH 689
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,777,676
Number of Sequences: 36976
Number of extensions: 120502
Number of successful extensions: 603
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 595
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 597
length of query: 285
length of database: 9,014,727
effective HSP length: 95
effective length of query: 190
effective length of database: 5,502,007
effective search space: 1045381330
effective search space used: 1045381330
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146940.1


