

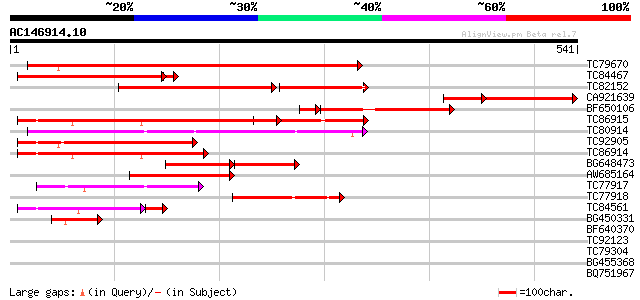
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146914.10 + phase: 0
(541 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC79670 similar to PIR|A84635|A84635 probable calmodulin-binding... 315 3e-86
TC84467 weakly similar to GP|10178088|dbj|BAB11507. contains sim... 277 8e-78
TC82152 similar to PIR|T03793|T03793 calmodulin-binding protein ... 200 2e-68
CA921639 184 2e-66
BF650106 similar to PIR|A84635|A846 probable calmodulin-binding ... 221 7e-66
TC86915 similar to PIR|D96765|D96765 hypothetical protein F25P22... 218 5e-57
TC80914 similar to PIR|T41098|T41098 WD-repeat protein - fission... 165 4e-41
TC92905 similar to GP|20197434|gb|AAD08944.2 putative calmodulin... 155 3e-38
TC86914 similar to GP|15528601|dbj|BAB64623. putative calmodulin... 154 1e-37
BG648473 weakly similar to GP|9758319|dbj calmodulin-binding pro... 137 1e-32
AW685164 similar to PIR|T03793|T03 calmodulin-binding protein TC... 115 4e-26
TC77917 weakly similar to PIR|D96765|D96765 hypothetical protein... 101 6e-22
TC77918 weakly similar to PIR|T03793|T03793 calmodulin-binding p... 91 1e-18
TC84561 similar to GP|20197434|gb|AAD08944.2 putative calmodulin... 73 7e-15
BG450331 similar to GP|16649103|gb| Unknown protein {Arabidopsis... 57 2e-08
BF640370 similar to GP|9758319|dbj| calmodulin-binding protein {... 36 0.033
TC92123 homologue to SP|Q42971|ENO_ORYSA Enolase (EC 4.2.1.11) (... 30 1.8
TC79304 similar to GP|9759186|dbj|BAB09801.1 emb|CAB66399.1~gene... 30 2.4
BG455368 weakly similar to GP|9759524|dbj selenium-binding prote... 29 5.2
BQ751967 weakly similar to GP|22293246|emb hexose transporter {K... 29 5.2
>TC79670 similar to PIR|A84635|A84635 probable calmodulin-binding protein
[imported] - Arabidopsis thaliana, partial (58%)
Length = 1213
Score = 315 bits (807), Expect = 3e-86
Identities = 156/323 (48%), Positives = 228/323 (70%), Gaps = 4/323 (1%)
Frame = +1
Query: 18 SVRNLMEPILEPLVRRVVREEVELALKK--HLSSIKQTCGKEMNTSESRTLQLQFENSIS 75
+ + L+ + EPL+R++V EEVE AL K H ++ + + LQL F +
Sbjct: 109 TAKTLLLHLWEPLLRKIVSEEVERALAKLDHAKLGDRSSPARIEAPGEKNLQLHFRTRMP 288
Query: 76 LPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEE-SDIWMPE 134
+FTG ++EGE G+ + + L+D TG VV GPES AK+ +VVLEGDF EE D W E
Sbjct: 289 PHLFTGGKVEGEQGAVIHVVLLDPNTGNVVQVGPESVAKLNVVVLEGDFNEEIDDDWTKE 468
Query: 135 DFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLGVRVVDNF- 193
F+++ V+ER+GK+PLLTGD+ + LK+G+ +G++S+TDNSSW RSR+FRLGV+V +
Sbjct: 469 HFESHEVKEREGKRPLLTGDLQVSLKEGVGTLGDLSFTDNSSWIRSRKFRLGVKVAPGYC 648
Query: 194 DGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVK 253
DGIR+RE KT++F V+DHRGELYKKH+PP+L DEVWRL++I KDGA H++L + +I TV+
Sbjct: 649 DGIRVREGKTEAFAVKDHRGELYKKHYPPALHDEVWRLDRIAKDGALHKKLIQSRIVTVE 828
Query: 254 DFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFNAV 313
DFL LL +P KLR++LG+GMS +MWE TVEHA+TCVL T V + + ++FN +
Sbjct: 829 DFLRLLVREPQKLRSVLGSGMSNRMWENTVEHAKTCVLGTKLFVYYTDKTHSTGIMFNNI 1008
Query: 314 GEVTGLLAESEYVAVDKLSETEK 336
E+ GL+A+ ++ +++ L+ +K
Sbjct: 1009YELRGLIADGQFFSLESLTSNQK 1077
>TC84467 weakly similar to GP|10178088|dbj|BAB11507. contains similarity to
calmodulin-binding protein~gene_id:K19B1.18 {Arabidopsis
thaliana}, partial (18%)
Length = 880
Score = 277 bits (709), Expect(2) = 8e-78
Identities = 142/142 (100%), Positives = 142/142 (100%)
Frame = +2
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSIKQTCGKEMNTSESRTLQ 67
SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSIKQTCGKEMNTSESRTLQ
Sbjct: 416 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSIKQTCGKEMNTSESRTLQ 595
Query: 68 LQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEE 127
LQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEE
Sbjct: 596 LQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEE 775
Query: 128 SDIWMPEDFKNNIVRERDGKKP 149
SDIWMPEDFKNNIVRERDGKKP
Sbjct: 776 SDIWMPEDFKNNIVRERDGKKP 841
Score = 32.0 bits (71), Expect(2) = 8e-78
Identities = 14/15 (93%), Positives = 14/15 (93%)
Frame = +3
Query: 147 KKPLLTGDVILYLKD 161
K PLLTGDVILYLKD
Sbjct: 834 KSPLLTGDVILYLKD 878
>TC82152 similar to PIR|T03793|T03793 calmodulin-binding protein TCB60 -
common tobacco, partial (42%)
Length = 876
Score = 200 bits (508), Expect(2) = 2e-68
Identities = 94/152 (61%), Positives = 127/152 (82%), Gaps = 2/152 (1%)
Frame = +1
Query: 105 VCTGPESSAKVEIVVLEGDFEEESDI-WMPEDFKNNIVRERDGKKPLLTGDVILYLKDGL 163
V +GPES AK+++ VLEGDF E D W E+F+++IV+ER+GK+PLL+GD+ + LKDG+
Sbjct: 1 VTSGPESCAKLDVFVLEGDFNNEDDENWSEEEFESHIVKEREGKRPLLSGDLQVILKDGV 180
Query: 164 CMVGEISYTDNSSWTRSRRFRLGVRVVDNF-DGIRIREAKTDSFIVRDHRGELYKKHHPP 222
+GEIS+TDNSSW RSR+FRLG++V F DG+RIREAK+++F V+DHRGELYKKH+PP
Sbjct: 181 GTLGEISFTDNSSWIRSRKFRLGLKVSSGFCDGMRIREAKSEAFTVKDHRGELYKKHYPP 360
Query: 223 SLSDEVWRLEKIGKDGAFHRRLSREKIRTVKD 254
+L DEVWRLEKIGKDG+FH+RL++ + V+D
Sbjct: 361 ALHDEVWRLEKIGKDGSFHKRLNKAGVFNVED 456
Score = 77.8 bits (190), Expect(2) = 2e-68
Identities = 38/85 (44%), Positives = 57/85 (66%)
Frame = +3
Query: 258 LLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFNAVGEVT 317
L++ P +L ILGTGMS KMW++ VEHA+TCVL +V + ++ VVFN + E++
Sbjct: 468 LVSETPQRLPNILGTGMSNKMWDIPVEHAKTCVLSGKLYVYYPEDARNVGVVFNHIYELS 647
Query: 318 GLLAESEYVAVDKLSETEKAFISDH 342
GL+A +Y D LSE++K +S+H
Sbjct: 648 GLIANDQYHTADSLSESQK--VSEH 716
>CA921639
Length = 683
Score = 184 bits (466), Expect(2) = 2e-66
Identities = 89/90 (98%), Positives = 89/90 (98%)
Frame = -1
Query: 452 LGFPVQVSNSLICDMDSIVHAFGDEDHLQFFDADLQSQCHIEADLHSAVDSFMPVSSTSM 511
L FPVQVSNSLICDMDSIVHAFGDEDHLQFFDADLQSQCHIEADLHSAVDSFMPVSSTSM
Sbjct: 569 LVFPVQVSNSLICDMDSIVHAFGDEDHLQFFDADLQSQCHIEADLHSAVDSFMPVSSTSM 390
Query: 512 TKGKAQRRWRKVVNVLKWFMVKKRRNQLYR 541
TKGKAQRRWRKVVNVLKWFMVKKRRNQLYR
Sbjct: 389 TKGKAQRRWRKVVNVLKWFMVKKRRNQLYR 300
Score = 87.8 bits (216), Expect(2) = 2e-66
Identities = 41/41 (100%), Positives = 41/41 (100%)
Frame = -3
Query: 415 ASSTDIMPSIYSVGGTSSLDDYGLPNFESLGLRYDQHLGFP 455
ASSTDIMPSIYSVGGTSSLDDYGLPNFESLGLRYDQHLGFP
Sbjct: 681 ASSTDIMPSIYSVGGTSSLDDYGLPNFESLGLRYDQHLGFP 559
>BF650106 similar to PIR|A84635|A846 probable calmodulin-binding protein
[imported] - Arabidopsis thaliana, partial (2%)
Length = 415
Score = 221 bits (564), Expect(2) = 7e-66
Identities = 117/128 (91%), Positives = 117/128 (91%)
Frame = +1
Query: 297 VSFASHSQQPHVVFNAVGEVTGLLAESEYVAVDKLSETEKAFISDHPLVSLADAQISVIS 356
VSFASHSQQPHVVFNAVGEVTGLLAESEYVAVDKLSETEKA DAQISVIS
Sbjct: 64 VSFASHSQQPHVVFNAVGEVTGLLAESEYVAVDKLSETEKA-----------DAQISVIS 210
Query: 357 ALNQCDFASFEDEVSLMDGYSHLTNVHYSPSSPRTEGSSANKLLALQKTGGFNYTQESAS 416
ALNQCDFASFEDEVSLMDGYSHLTNVHYSPSSPRTEGSSANKLLALQKTGGFNYTQESAS
Sbjct: 211 ALNQCDFASFEDEVSLMDGYSHLTNVHYSPSSPRTEGSSANKLLALQKTGGFNYTQESAS 390
Query: 417 STDIMPSI 424
STDIMPSI
Sbjct: 391 STDIMPSI 414
Score = 47.8 bits (112), Expect(2) = 7e-66
Identities = 20/20 (100%), Positives = 20/20 (100%)
Frame = +3
Query: 277 KMWEVTVEHARTCVLDTTRH 296
KMWEVTVEHARTCVLDTTRH
Sbjct: 3 KMWEVTVEHARTCVLDTTRH 62
>TC86915 similar to PIR|D96765|D96765 hypothetical protein F25P22.22
[imported] - Arabidopsis thaliana, partial (38%)
Length = 1635
Score = 218 bits (555), Expect = 5e-57
Identities = 113/259 (43%), Positives = 170/259 (65%), Gaps = 7/259 (2%)
Frame = +2
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSIKQTCGKEM----NTSES 63
SV+ E++ +++++NL LEPL+RRVV EEVE A+++ ++ E
Sbjct: 149 SVIGEIVMVKNMQNLFSG-LEPLLRRVVNEEVERAMRQCYPPVRSITNSPSLRLKAMEEP 325
Query: 64 RTLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGD 123
+ Q F+ +SLP+FTG+RI DG+++ + LVD ++V T K+EIVVL+GD
Sbjct: 326 LSFQFMFKKKLSLPIFTGSRILDMDGNSINVILVDKSNDQIVPTSLPHPIKIEIVVLDGD 505
Query: 124 F--EEESDIWMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSR 181
F E+ W E+F +NIV+ER+GK+PLLTG++ L ++DG+ +G+I +TDNSSW RSR
Sbjct: 506 FPPSEKESSWTSEEFNSNIVKERNGKRPLLTGELNLTMRDGIAPIGDIEFTDNSSWIRSR 685
Query: 182 RFRLGVRVVDNFD-GIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAF 240
+FR+ VR+ + +RI E T+ F+V+DHRGELYKKHHPPSL+DEVWRLEK+GK
Sbjct: 686 KFRVAVRIAPGTNQSVRILEGMTEPFVVKDHRGELYKKHHPPSLNDEVWRLEKLGKMELS 865
Query: 241 HRRLSREKIRTVKDFLTLL 259
+ ++ ++ K F + L
Sbjct: 866 TKN*PQKGLQQFKSFSSCL 922
Score = 81.6 bits (200), Expect = 7e-16
Identities = 41/110 (37%), Positives = 68/110 (61%)
Frame = +1
Query: 233 KIGKDGAFHRRLSREKIRTVKDFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLD 292
+IGKDGAFH++L+ + I TV++FL L +DP +LR ILG GMS KMW+VT++HA+TCV+
Sbjct: 841 EIGKDGAFHKKLTSKGITTVQEFLKLSVVDPLRLRKILGIGMSEKMWDVTIKHAKTCVMG 1020
Query: 293 TTRHVSFASHSQQPHVVFNAVGEVTGLLAESEYVAVDKLSETEKAFISDH 342
+V Q + NA+ ++ + + + +K++I ++
Sbjct: 1021NKLYV---YRGPQFTIHLNAICQMVRANINGQTIPNRDICNMDKSYIQNY 1161
>TC80914 similar to PIR|T41098|T41098 WD-repeat protein - fission yeast
(Schizosaccharomyces pombe), partial (4%)
Length = 1576
Score = 165 bits (418), Expect = 4e-41
Identities = 115/331 (34%), Positives = 182/331 (54%), Gaps = 7/331 (2%)
Frame = -1
Query: 18 SVRNLMEPILEPLVRRVVREEVELALKKHL-SSIKQTCGKEMNTSESRT-LQLQFENSIS 75
+ RN EPL+R+V++EE++ L +L S T + NT E R LQL F N +
Sbjct: 1315 AARNTFAFYFEPLLRKVMQEEIQHKLHHYLIPSCGLTLNELANTYERRRRLQLCFMNKMP 1136
Query: 76 LPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEESDIWMPED 135
+FT + I+ E L+I L DA + + SS K+++ VL GDFE+E W E+
Sbjct: 1135 ETIFTMSEIKAEGDEPLQIELRDAGNXQRIVREEGSSMKIQLCVLYGDFEKED--WTAEE 962
Query: 136 FKNNIVRERDGKKPLLTGDVILYLKDGLCMVG-EISYTDNSSWTRSRRFRLGVRVVD-NF 193
F IV R GK LL G+ I+ L++G+ + EI +TD S R+ +FRLGV++V N
Sbjct: 961 FNTQIVPPRVGKGQLLKGNKIITLRNGVADINIEIEFTDISK-RRTGQFRLGVKIVQSNS 785
Query: 194 DGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVK 253
G IRE +++ F V D RG+ Y+K PSL+D+VWRL I +DG ++L+ + I T+K
Sbjct: 784 IGACIREGRSEPFKVLDVRGKAYEKRDRPSLNDKVWRLRGIRRDGTLDKQLALDGIHTIK 605
Query: 254 DFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTC-VLDTTRHVSFASHSQQPHVVFNA 312
DFL LL + LR + K+W+ + HA+ C V D ++ + + +VFN+
Sbjct: 604 DFLQLLITNEDSLRNKIDK-TGGKLWKKIIAHAKHCDVDDDGCYIYYVTEQPISSLVFNS 428
Query: 313 VGEVTGLLAESEY--VAVDKLSETEKAFISD 341
+ +V + ++ ++ L+ EK + +
Sbjct: 427 ILKVVEVTFHNQQNSRSIQSLNPQEKRLVEE 335
>TC92905 similar to GP|20197434|gb|AAD08944.2 putative calmodulin-binding
protein {Arabidopsis thaliana}, partial (55%)
Length = 815
Score = 155 bits (393), Expect = 3e-38
Identities = 82/177 (46%), Positives = 123/177 (69%), Gaps = 5/177 (2%)
Frame = +3
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKK----HLSSIKQTCGKEMNTSES 63
SV+ E +K+ S++ L LEP++RRVV EEVE AL K +LS ++ K + +
Sbjct: 294 SVIVEALKVDSLQKLCSS-LEPILRRVVSEEVERALAKLAPTNLSG--RSSPKRIENPDG 464
Query: 64 RTLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGD 123
LQL+F +SLP+FTG ++EGE G+ + I L+DA TG VV +GP S +++++VLEGD
Sbjct: 465 GNLQLKFRTRLSLPLFTGGKVEGEQGTAIHIVLIDANTGHVVTSGPASCVRLDVIVLEGD 644
Query: 124 F-EEESDIWMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTR 179
F E+ D W E+F ++IV+ER+GK+PLLTGD+ + LKDG+ + ++++TDNSSW R
Sbjct: 645 FNNEDDDTWSQEEFDSHIVKEREGKRPLLTGDLQVTLKDGVGTLADLTFTDNSSWIR 815
>TC86914 similar to GP|15528601|dbj|BAB64623. putative calmodulin-binding
protein {Oryza sativa (japonica cultivar-group)},
partial (18%)
Length = 727
Score = 154 bits (388), Expect = 1e-37
Identities = 81/188 (43%), Positives = 124/188 (65%), Gaps = 6/188 (3%)
Frame = +2
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKKHLSSIKQTCGKEM----NTSES 63
SV+ EV+ +++++NL LEPL+RRVV EEVE A+++ ++ E
Sbjct: 164 SVIGEVVMVKNMQNLFSG-LEPLLRRVVNEEVERAMRQCYPPVRSITKSPSLRLEGMEEP 340
Query: 64 RTLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGD 123
+LQ F+N +SLP+FTG++I DG+++ + LVD +VV T K+EIVVL+GD
Sbjct: 341 LSLQFMFKNKLSLPIFTGSKILDMDGNSISVILVDRSNDQVVPTSLSHPIKIEIVVLDGD 520
Query: 124 F--EEESDIWMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSR 181
F ++ W E+F N+IV+ER GK+PLLTG++ L ++DG+ +G+I +TDNSSW RSR
Sbjct: 521 FPPSKKESSWTSEEFNNSIVKERTGKRPLLTGELNLTMRDGIAPIGDIEFTDNSSWIRSR 700
Query: 182 RFRLGVRV 189
+FR+ VR+
Sbjct: 701 KFRVAVRI 724
>BG648473 weakly similar to GP|9758319|dbj calmodulin-binding protein
{Arabidopsis thaliana}, partial (19%)
Length = 822
Score = 137 bits (344), Expect = 1e-32
Identities = 66/66 (100%), Positives = 66/66 (100%)
Frame = +2
Query: 149 PLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLGVRVVDNFDGIRIREAKTDSFIV 208
PLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLGVRVVDNFDGIRIREAKTDSFIV
Sbjct: 2 PLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRLGVRVVDNFDGIRIREAKTDSFIV 181
Query: 209 RDHRGE 214
RDHRGE
Sbjct: 182 RDHRGE 199
Score = 124 bits (311), Expect = 9e-29
Identities = 60/62 (96%), Positives = 61/62 (97%)
Frame = +2
Query: 215 LYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVKDFLTLLNLDPAKLRTILGTGM 274
+YK HHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVKDFLTLLNLDPAKLRTILGTGM
Sbjct: 635 VYKIHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVKDFLTLLNLDPAKLRTILGTGM 814
Query: 275 SA 276
SA
Sbjct: 815 SA 820
>AW685164 similar to PIR|T03793|T03 calmodulin-binding protein TCB60 - common
tobacco, partial (18%)
Length = 353
Score = 115 bits (288), Expect = 4e-26
Identities = 55/102 (53%), Positives = 79/102 (76%), Gaps = 2/102 (1%)
Frame = +1
Query: 115 VEIVVLEGDFEEESDI-WMPEDFKNNIVRERDGKKPLLTGDVILYLKDGLCMVGEISYTD 173
+++VVLEGDF E D W E+F++++V+ER GK+PLL G++ + LK+G+ +GE+ +TD
Sbjct: 1 LDVVVLEGDFNNEDDEDWSQEEFESHVVKERQGKRPLLNGELQVTLKEGVGTLGELIFTD 180
Query: 174 NSSWTRSRRFRLGVRVVDNF-DGIRIREAKTDSFIVRDHRGE 214
NSSW RSR+FRLG++V F + IRIREAKT +F V+DH GE
Sbjct: 181 NSSWIRSRKFRLGMKVASGFGESIRIREAKTVAFTVKDHXGE 306
>TC77917 weakly similar to PIR|D96765|D96765 hypothetical protein F25P22.22
[imported] - Arabidopsis thaliana, partial (10%)
Length = 716
Score = 101 bits (252), Expect = 6e-22
Identities = 60/164 (36%), Positives = 93/164 (56%), Gaps = 4/164 (2%)
Frame = +1
Query: 26 ILEPLVRRVVREEVELALKKHLSSIKQTCGKEMNTSESRTLQLQF----ENSISLPVFTG 81
+LE +R VVR+ VE ++ L S ++ E S +R L+L F N +S P F+
Sbjct: 235 LLESFIRGVVRDVVESKFQERLLSSEKA--NEAGKSGARPLELCFINNNNNKLSGPFFSQ 408
Query: 82 ARIEGEDGSNLRIRLVDALTGKVVCTGPESSAKVEIVVLEGDFEEESDIWMPEDFKNNIV 141
+ I +D L++ L D + +V GP SS K+EI L+G F E W +FK NI+
Sbjct: 409 SNIIAKDEPPLQVALFDVGSKSIVNVGPFSSTKIEICALDGGFGSED--WTEIEFKANIL 582
Query: 142 RERDGKKPLLTGDVILYLKDGLCMVGEISYTDNSSWTRSRRFRL 185
RERDGK+PLL G+ + LK+G+ + + +TDNS W + + ++
Sbjct: 583 RERDGKQPLLVGERFITLKNGVASISKTIFTDNSRWLKKQNVQV 714
>TC77918 weakly similar to PIR|T03793|T03793 calmodulin-binding protein
TCB60 - common tobacco, partial (7%)
Length = 1199
Score = 90.5 bits (223), Expect = 1e-18
Identities = 48/107 (44%), Positives = 68/107 (62%)
Frame = +1
Query: 213 GELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVKDFLTLLNLDPAKLRTILGT 272
G+ +KH PP L+D VWRLEKI KDG FH+RLS I TVKD L LL ++ + L I
Sbjct: 1 GQPNEKHFPPFLNDFVWRLEKIAKDGRFHKRLSSNGIHTVKDLLQLLIINESSLHGIF-E 177
Query: 273 GMSAKMWEVTVEHARTCVLDTTRHVSFASHSQQPHVVFNAVGEVTGL 319
+ K W +EHA++CVLD + S+ + QP ++FNA+ ++ G+
Sbjct: 178 KIQRKSWLAIIEHAKSCVLDDHKLYSYGTIG-QPILLFNAIYKLVGV 315
>TC84561 similar to GP|20197434|gb|AAD08944.2 putative calmodulin-binding
protein {Arabidopsis thaliana}, partial (7%)
Length = 632
Score = 72.8 bits (177), Expect(2) = 7e-15
Identities = 48/126 (38%), Positives = 74/126 (58%), Gaps = 4/126 (3%)
Frame = +1
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKK-HLSSIKQTCGKEMNTSESR-- 64
SV+ +K+Q+++N + LEP ++RVV EEV+ A+ K SSI ++ + +
Sbjct: 166 SVIETAVKVQNMQNFLGA-LEPFLKRVVSEEVDRAIGKCSPSSINRSPSLRIQAPRDQQP 342
Query: 65 TLQLQFENSISLPVFTGARIEGEDGSNLRIRLVDAL-TGKVVCTGPESSAKVEIVVLEGD 123
TLQL F +SLP+FTG+RI +G + I LVD ++V T K+EIVVL+GD
Sbjct: 343 TLQLSFSKRLSLPIFTGSRILDAEGKPICISLVDKTNNNQIVQTSLPYPIKLEIVVLDGD 522
Query: 124 FEEESD 129
F + +
Sbjct: 523 FPHDEN 540
Score = 25.8 bits (55), Expect(2) = 7e-15
Identities = 9/21 (42%), Positives = 15/21 (70%)
Frame = +2
Query: 130 IWMPEDFKNNIVRERDGKKPL 150
IW E+F +IV+ER G++ +
Sbjct: 545 IWTSEEFNKDIVKERTGEETI 607
>BG450331 similar to GP|16649103|gb| Unknown protein {Arabidopsis thaliana},
partial (12%)
Length = 433
Score = 57.0 bits (136), Expect = 2e-08
Identities = 31/52 (59%), Positives = 38/52 (72%), Gaps = 4/52 (7%)
Frame = +3
Query: 41 LALKKHLSSIK----QTCGKEMNTSESRTLQLQFENSISLPVFTGARIEGED 88
L L HL ++ ++CGK M+TS SRTLQLQF SIS+PVFT ARIEGE+
Sbjct: 114 LCLSTHL*NLNMLHYRSCGKAMDTS*SRTLQLQFGKSISIPVFTRARIEGEE 269
>BF640370 similar to GP|9758319|dbj| calmodulin-binding protein {Arabidopsis
thaliana}, partial (13%)
Length = 521
Score = 36.2 bits (82), Expect = 0.033
Identities = 20/38 (52%), Positives = 27/38 (70%)
Frame = +1
Query: 8 SVVREVMKLQSVRNLMEPILEPLVRRVVREEVELALKK 45
SV+ E +K+ S++ L LEP++RRVV EEVE AL K
Sbjct: 295 SVIVEALKVDSMQKLCSS-LEPILRRVVSEEVERALAK 405
>TC92123 homologue to SP|Q42971|ENO_ORYSA Enolase (EC 4.2.1.11)
(2-phosphoglycerate dehydratase) (2-phospho-D- glycerate
hydro-lyase) (OSE1)., partial (48%)
Length = 990
Score = 30.4 bits (67), Expect = 1.8
Identities = 12/27 (44%), Positives = 19/27 (69%)
Frame = +1
Query: 323 SEYVAVDKLSETEKAFISDHPLVSLAD 349
SE ++ D L K+F+SD+P+VS+ D
Sbjct: 139 SEKISGDSLKNVYKSFVSDYPIVSIED 219
>TC79304 similar to GP|9759186|dbj|BAB09801.1
emb|CAB66399.1~gene_id:K19E1.19~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (8%)
Length = 790
Score = 30.0 bits (66), Expect = 2.4
Identities = 24/103 (23%), Positives = 42/103 (40%)
Frame = +2
Query: 194 DGIRIREAKTDSFIVRDHRGELYKKHHPPSLSDEVWRLEKIGKDGAFHRRLSREKIRTVK 253
+ + ++ + S +VRDHRG E WR KI D ++ E + K
Sbjct: 155 NSVMLQHPRFTSLMVRDHRGV------------EHWRPIKIDLDRHIFI-INNESVSEDK 295
Query: 254 DFLTLLNLDPAKLRTILGTGMSAKMWEVTVEHARTCVLDTTRH 296
D + ++ + L T M +WE+ + A C++ H
Sbjct: 296 DDESAISEYMSDLCTESKLSMDKPLWEMHILRAHKCIIFRIHH 424
>BG455368 weakly similar to GP|9759524|dbj selenium-binding protein-like
{Arabidopsis thaliana}, partial (33%)
Length = 687
Score = 28.9 bits (63), Expect = 5.2
Identities = 16/41 (39%), Positives = 23/41 (56%), Gaps = 1/41 (2%)
Frame = -2
Query: 389 PRTEGSSANKLL-ALQKTGGFNYTQESASSTDIMPSIYSVG 428
PR GSS+NKL A+ T GF + ++ S P + S+G
Sbjct: 161 PRLSGSSSNKLREAISATSGFPWMCDAPRSAPHTPPLGSIG 39
>BQ751967 weakly similar to GP|22293246|emb hexose transporter {Kluyveromyces
lactis}, partial (17%)
Length = 492
Score = 28.9 bits (63), Expect = 5.2
Identities = 22/80 (27%), Positives = 38/80 (47%), Gaps = 2/80 (2%)
Frame = -1
Query: 25 PILEPLVRR--VVREEVELALKKHLSSIKQTCGKEMNTSESRTLQLQFENSISLPVFTGA 82
P + +VR VVR + + L +HL ++ G + + S R L+ + + LPV A
Sbjct: 447 PHVPQIVRHDHVVRPALPVLLHRHLQLLRHAGGHDPHRSHPRLLRRHRHSRMVLPVALHA 268
Query: 83 RIEGEDGSNLRIRLVDALTG 102
+G+D + L +A G
Sbjct: 267 *DQGQDPGRH*LALREAHPG 208
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,919,166
Number of Sequences: 36976
Number of extensions: 204762
Number of successful extensions: 1076
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 1049
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1053
length of query: 541
length of database: 9,014,727
effective HSP length: 101
effective length of query: 440
effective length of database: 5,280,151
effective search space: 2323266440
effective search space used: 2323266440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146914.10


