

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
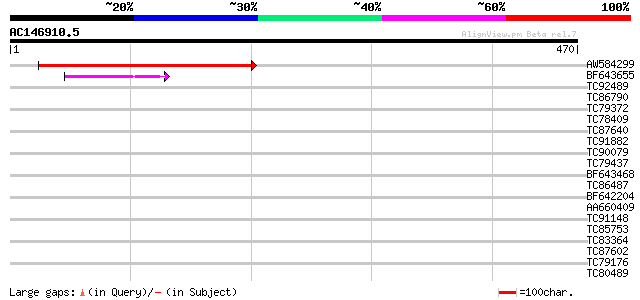
Query= AC146910.5 - phase: 0 /pseudo
(470 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW584299 similar to GP|20160694|db hypothetical protein~similar ... 365 e-101
BF643655 similar to GP|6056190|gb| putative glycerol-3-phosphate... 42 4e-04
TC92489 homologue to GP|13274996|emb|CAC34013. soybean protox-1 ... 35 0.062
TC86790 similar to PIR|A84861|A84861 probable amine oxidase [imp... 34 0.11
TC79372 homologue to GP|2281647|gb|AAC49777.1| AP2 domain contai... 33 0.31
TC78409 homologue to GP|18086418|gb|AAL57665.1 AT3g59050/F17J16_... 33 0.31
TC87640 similar to PIR|C96618|C96618 probable squalene monooxyge... 32 0.40
TC91882 similar to GP|10176714|dbj|BAB09944. dimethylaniline mon... 32 0.68
TC90079 similar to GP|5454204|gb|AAD43619.1| T3P18.18 {Arabidops... 31 1.2
TC79437 similar to PIR|G96720|G96720 nitrate transporter (NTL1) ... 30 1.5
BF643468 weakly similar to PIR|F96682|F96 hypothetical protein F... 30 1.5
TC86487 similar to PIR|C84473|C84473 probable protein kinase [im... 30 2.6
BF642204 similar to GP|6056190|gb| putative glycerol-3-phosphate... 30 2.6
AA660409 weakly similar to GP|22655529|gb| water-stress protein ... 29 3.4
TC91148 similar to GP|5732036|gb|AAD48937.1| similar to disease ... 29 3.4
TC85753 similar to PIR|D96682|D96682 protein F1E22.18 [imported]... 29 4.4
TC83364 similar to GP|13786442|gb|AAK39567.1 putative electron t... 29 4.4
TC87602 similar to GP|21741453|emb|CAD41397. OJ000223_09.9 {Oryz... 28 5.8
TC79176 similar to GP|20259085|gb|AAM14258.1 putative argininosu... 28 5.8
TC80489 similar to GP|9294341|dbj|BAB02238.1 contains similarity... 28 5.8
>AW584299 similar to GP|20160694|db hypothetical protein~similar to
Arabidopsis thaliana chromosome 3 At3g56840, partial
(22%)
Length = 549
Score = 365 bits (937), Expect = e-101
Identities = 179/180 (99%), Positives = 179/180 (99%)
Frame = +2
Query: 25 VHMKWNNPFNNWVRNIRSTTQNDTNTTTSYPVSRETTTSYSVPRERVDCVVIGAGVVGIA 84
VHMKWNNPFNNWVRNIRSTTQNDTNTTTSYPVSRETTTSYSVPRERVDCVVIGAGVVGIA
Sbjct: 8 VHMKWNNPFNNWVRNIRSTTQNDTNTTTSYPVSRETTTSYSVPRERVDCVVIGAGVVGIA 187
Query: 85 VARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSLKAIFCVKGREMLYEYC 144
VARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSLKAIFCVKGREMLYEYC
Sbjct: 188 VARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSLKAIFCVKGREMLYEYC 367
Query: 145 AKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMDGVDAMKMEPELQCVKA 204
AKHDIPHEQTGKLIVATRSSEIPKLSVILNHGI NGVDGLKMMDGVDAMKMEPELQCVKA
Sbjct: 368 AKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIHNGVDGLKMMDGVDAMKMEPELQCVKA 547
>BF643655 similar to GP|6056190|gb| putative glycerol-3-phosphate
dehydrogenase {Arabidopsis thaliana}, partial (27%)
Length = 587
Score = 42.4 bits (98), Expect = 4e-04
Identities = 27/87 (31%), Positives = 44/87 (50%)
Frame = +3
Query: 46 NDTNTTTSYPVSRETTTSYSVPRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSF 105
+D N ++++ S +D +VIG G G VA +G V ++E F
Sbjct: 60 HDPNAVLPSRQVQQSSLIGSTSANPLDILVIGGGATGTGVALDATTRGLRVGLVERE-DF 236
Query: 106 GTGTSSRNSEVVHAGIYYPHHSLKAIF 132
+GTSSR++++VH G+ Y KA+F
Sbjct: 237 SSGTSSRSTKLVHGGVRYLE---KAVF 308
>TC92489 homologue to GP|13274996|emb|CAC34013. soybean protox-1 {Glycine
max}, partial (23%)
Length = 635
Score = 35.0 bits (79), Expect = 0.062
Identities = 15/36 (41%), Positives = 24/36 (66%)
Frame = +1
Query: 57 SRETTTSYSVPRERVDCVVIGAGVVGIAVARALALK 92
++ T+ S S VDCVV+G G+ G+ +A+AL+ K
Sbjct: 226 TKTTSKSNSGESSSVDCVVVGGGISGLCIAQALSTK 333
>TC86790 similar to PIR|A84861|A84861 probable amine oxidase [imported] -
Arabidopsis thaliana, partial (91%)
Length = 2198
Score = 34.3 bits (77), Expect = 0.11
Identities = 17/40 (42%), Positives = 23/40 (57%)
Frame = +2
Query: 67 PRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFG 106
PR +VIG G+ GIA ARAL +V+++ES G
Sbjct: 449 PRRSPSVIVIGGGMAGIAAARALHDASFQVVLLESRDRIG 568
>TC79372 homologue to GP|2281647|gb|AAC49777.1| AP2 domain containing
protein RAP2.11 {Arabidopsis thaliana}, partial (27%)
Length = 1086
Score = 32.7 bits (73), Expect = 0.31
Identities = 26/111 (23%), Positives = 47/111 (41%), Gaps = 11/111 (9%)
Frame = +3
Query: 17 SSTRTRDHVHMKWNNPFNNWVRNI-----RSTTQNDTN---TTTSYPVSRETTTSYSVPR 68
S TRT H+ +++P + +R++ + T Q D N +TTS+ + TTS
Sbjct: 168 SKTRTNFVTHVSYDSPLASRIRHLLNNRKKGTKQQDMNGISSTTSHADTTNDTTSDGSTS 347
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIES---APSFGTGTSSRNSEV 116
+C+ +G + A + + S + S T+S N+ V
Sbjct: 348 STTNCIGTASGAINSTSASGVTSTSTNISTSASGVASTSTDISTNSSNTNV 500
>TC78409 homologue to GP|18086418|gb|AAL57665.1 AT3g59050/F17J16_100
{Arabidopsis thaliana}, partial (19%)
Length = 689
Score = 32.7 bits (73), Expect = 0.31
Identities = 17/42 (40%), Positives = 23/42 (54%)
Frame = +1
Query: 65 SVPRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFG 106
SV +VIG G+ GIA ARAL +V+++ES G
Sbjct: 382 SVQERSPSVIVIGGGMAGIAAARALQDASFQVVLLESRERLG 507
>TC87640 similar to PIR|C96618|C96618 probable squalene monooxygenase
F9K23.3 [imported] - Arabidopsis thaliana, partial (55%)
Length = 1288
Score = 32.3 bits (72), Expect = 0.40
Identities = 15/29 (51%), Positives = 19/29 (64%)
Frame = +1
Query: 72 DCVVIGAGVVGIAVARALALKGREVIVIE 100
D +++GAGV G A+A L GR V VIE
Sbjct: 385 DIIIVGAGVAGAALAYTLGKDGRRVHVIE 471
>TC91882 similar to GP|10176714|dbj|BAB09944. dimethylaniline
monooxygenase-like protein {Arabidopsis thaliana},
partial (47%)
Length = 800
Score = 31.6 bits (70), Expect = 0.68
Identities = 18/53 (33%), Positives = 27/53 (49%)
Frame = +2
Query: 54 YPVSRETTTSYSVPRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFG 106
Y +S T S + + + VIGAG G+ AR + +G +VIV+E G
Sbjct: 5 YFLSLATMVSETNQYQSKNVCVIGAGPAGLVAAREIRKEGHKVIVLEQNHDIG 163
>TC90079 similar to GP|5454204|gb|AAD43619.1| T3P18.18 {Arabidopsis
thaliana}, partial (38%)
Length = 677
Score = 30.8 bits (68), Expect = 1.2
Identities = 23/64 (35%), Positives = 29/64 (44%), Gaps = 12/64 (18%)
Frame = +3
Query: 71 VDCVVIGAGVVGIAVARALALKGREVIVIESAPSFG---TGTSSRNS---------EVVH 118
V VIGAGV G+ A+ L +G VIV E G TS +S E VH
Sbjct: 15 VKVAVIGAGVSGMVAAKELQQEGHNVIVFEKNNRVGGTWIYTSKSDSDPLSIDPTRETVH 194
Query: 119 AGIY 122
+ +Y
Sbjct: 195 SSVY 206
>TC79437 similar to PIR|G96720|G96720 nitrate transporter (NTL1)
53025-56402 [imported] - Arabidopsis thaliana, partial
(61%)
Length = 1424
Score = 30.4 bits (67), Expect = 1.5
Identities = 21/70 (30%), Positives = 36/70 (51%), Gaps = 7/70 (10%)
Frame = +3
Query: 9 LEQFKSSSSSTRTRDHVHMKWNNPFNNWVRNIRSTTQNDTNTTTSY------PVSRETTT 62
++ FKS +S RTR + K N+ +N +R + + Q++ T S+ P +R T
Sbjct: 867 IKPFKSKHTSKRTRTTSNPKTNHTISNTIRISKISQQSNHKQTNSFITTMHNPTTRRCQT 1046
Query: 63 S-YSVPRERV 71
S ++P RV
Sbjct: 1047SLQNIPDFRV 1076
>BF643468 weakly similar to PIR|F96682|F96 hypothetical protein F12P19.2
[imported] - Arabidopsis thaliana, partial (33%)
Length = 655
Score = 30.4 bits (67), Expect = 1.5
Identities = 23/71 (32%), Positives = 32/71 (44%), Gaps = 12/71 (16%)
Frame = +2
Query: 71 VDCVVIGAGVVGIAVARALALKGREVIVIES----------APSFGTGTSSRN--SEVVH 118
V VIGAGV G+ A+ L +G V+V E P + S + E VH
Sbjct: 83 VKVAVIGAGVSGLVAAKELQQEGHNVVVFEKNNRVGGTWIYTPKTDSDPLSMDPARETVH 262
Query: 119 AGIYYPHHSLK 129
+ +Y HSL+
Sbjct: 263 SSVY---HSLR 286
>TC86487 similar to PIR|C84473|C84473 probable protein kinase [imported] -
Arabidopsis thaliana, partial (75%)
Length = 2229
Score = 29.6 bits (65), Expect = 2.6
Identities = 18/66 (27%), Positives = 30/66 (45%)
Frame = +1
Query: 5 TIQTLEQFKSSSSSTRTRDHVHMKWNNPFNNWVRNIRSTTQNDTNTTTSYPVSRETTTSY 64
TI+T ++ K SSS+ T H H + N NN + N+ N + ++ Y
Sbjct: 1582 TIETPKERKRESSSSSTSTHHHHRRNQDVNNSPKPKHLIIDNNNNEVLVH--KKDGEEYY 1755
Query: 65 SVPRER 70
P+E+
Sbjct: 1756 DTPKEK 1773
>BF642204 similar to GP|6056190|gb| putative glycerol-3-phosphate
dehydrogenase {Arabidopsis thaliana}, partial (25%)
Length = 481
Score = 29.6 bits (65), Expect = 2.6
Identities = 19/56 (33%), Positives = 29/56 (50%)
Frame = +2
Query: 77 GAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSLKAIF 132
GAG AV R L + + F +GTSSR+++++H G+ Y KA+F
Sbjct: 8 GAGAALDAVTRGLK------VGLVEREDFASGTSSRSTKLLHGGVRYLE---KAVF 148
>AA660409 weakly similar to GP|22655529|gb| water-stress protein {Zea mays},
partial (16%)
Length = 644
Score = 29.3 bits (64), Expect = 3.4
Identities = 16/46 (34%), Positives = 23/46 (49%), Gaps = 7/46 (15%)
Frame = -1
Query: 32 PFNNWVRNIRSTTQN-------DTNTTTSYPVSRETTTSYSVPRER 70
P +NW R+ R++ + T + +SYP RETT S S R
Sbjct: 269 PTSNWYRHDRASISSXNRPDWRSTTSNSSYPTRRETTRSRSXAPRR 132
>TC91148 similar to GP|5732036|gb|AAD48937.1| similar to disease resistance
proteins; contains similarity ot Pfam family PF00560 -
Leucine Rich, partial (4%)
Length = 630
Score = 29.3 bits (64), Expect = 3.4
Identities = 11/28 (39%), Positives = 19/28 (67%)
Frame = +3
Query: 27 MKWNNPFNNWVRNIRSTTQNDTNTTTSY 54
+KW + F+N +++ ST +N TNT T +
Sbjct: 156 LKWKHSFDNQSQSLLSTWKNTTNTCTKW 239
>TC85753 similar to PIR|D96682|D96682 protein F1E22.18 [imported] -
Arabidopsis thaliana, partial (85%)
Length = 2024
Score = 28.9 bits (63), Expect = 4.4
Identities = 16/42 (38%), Positives = 22/42 (52%)
Frame = +1
Query: 65 SVPRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFG 106
S R +V+GAG+ GIA AR L +V ++ES G
Sbjct: 445 SQQRPAPSVIVVGAGISGIAAARILHDASFKVTLLESRDRLG 570
>TC83364 similar to GP|13786442|gb|AAK39567.1 putative electron transfer
oxidoreductase {Oryza sativa}, partial (11%)
Length = 381
Score = 28.9 bits (63), Expect = 4.4
Identities = 21/72 (29%), Positives = 30/72 (41%), Gaps = 6/72 (8%)
Frame = +2
Query: 48 TNTTTSYPVSRETTTSYSVPRERVDCVVIGAGVVGIAVARALALKGRE------VIVIES 101
T+ + P+SR T+ D V++GAG G++ A L RE V V+E
Sbjct: 125 THANSRVPISRNYCTAPERDSIEYDVVIVGAGPAGLSAAIRLKQLCRENDTDLSVCVLEK 304
Query: 102 APSFGTGTSSRN 113
G S N
Sbjct: 305 GSEVGAHILSGN 340
>TC87602 similar to GP|21741453|emb|CAD41397. OJ000223_09.9 {Oryza sativa},
partial (95%)
Length = 1263
Score = 28.5 bits (62), Expect = 5.8
Identities = 25/95 (26%), Positives = 37/95 (38%), Gaps = 9/95 (9%)
Frame = -1
Query: 127 SLKAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKM 186
SL IFC G AKHD+P + L +P+ ++ I N L
Sbjct: 1101 SLSIIFCKAGALTTQSLSAKHDLP--SSAALQSCGSKGTLPRKGTAISLHIFNAPPRLLS 928
Query: 187 MDGVDAMK---------MEPELQCVKAILSPLSGI 212
+ A+K M ++ V + +SPLS I
Sbjct: 927 NTSLQAIKFSISGAFSFMYSDIHSVTSFISPLSVI 823
>TC79176 similar to GP|20259085|gb|AAM14258.1 putative argininosuccinate
synthase {Arabidopsis thaliana}, partial (41%)
Length = 836
Score = 28.5 bits (62), Expect = 5.8
Identities = 28/111 (25%), Positives = 46/111 (41%), Gaps = 8/111 (7%)
Frame = +2
Query: 56 VSRETTTSYSVPRERVDCVVIGAGVVGIAVARALALKGREVIVIE-SAPSFGTGTSSRNS 114
+ E Y P R + ++G A+AR + K I E A + G + + +
Sbjct: 422 LKEEFVRDYVFPCLRAGAIYERKYLLGTAIARPVIAKAMVDIANEVGADAVSHGCTGKGN 601
Query: 115 EVVHAGIYY----PHHSLKAIFC---VKGREMLYEYCAKHDIPHEQTGKLI 158
+ V + + P ++ A + + GRE EY KHD+P T K I
Sbjct: 602 DQVRFELSFFALNPKLNVVAPWREWDITGREDAIEYAKKHDVPVPVTKKSI 754
>TC80489 similar to GP|9294341|dbj|BAB02238.1 contains similarity to unknown
protein~gb|AAF01552.1~gene_id:T6J22.18 {Arabidopsis
thaliana}, partial (68%)
Length = 1399
Score = 28.5 bits (62), Expect = 5.8
Identities = 13/25 (52%), Positives = 16/25 (64%)
Frame = +2
Query: 318 AYYARGCYFTLSNTKASPFRHLIYP 342
+YY GC FT S++ SP HLI P
Sbjct: 548 SYYRFGCSFTQSSSCHSPQSHLIRP 622
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,762,150
Number of Sequences: 36976
Number of extensions: 208566
Number of successful extensions: 1150
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 1134
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1146
length of query: 470
length of database: 9,014,727
effective HSP length: 100
effective length of query: 370
effective length of database: 5,317,127
effective search space: 1967336990
effective search space used: 1967336990
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146910.5


