

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
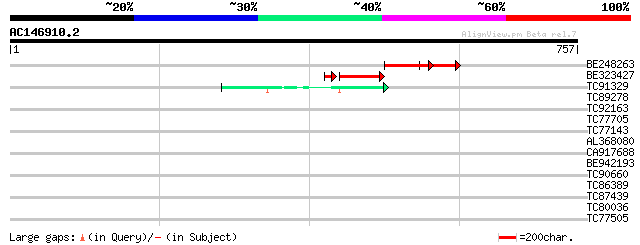
Query= AC146910.2 + phase: 1 /pseudo/partial
(757 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BE248263 similar to GP|17528976|gb| unknown protein {Arabidopsis... 135 8e-32
BE323427 similar to GP|17528976|gb| unknown protein {Arabidopsis... 107 7e-29
TC91329 similar to GP|7682783|gb|AAF67364.1| Hypothetical protei... 46 6e-05
TC89278 similar to GP|9757719|dbj|BAB08244.1 CRN (crooked neck) ... 42 0.001
TC92163 similar to PIR|C84861|C84861 hypothetical protein At2g43... 32 0.91
TC77705 similar to GP|21553857|gb|AAM62950.1 unknown {Arabidopsi... 31 1.5
TC77143 similar to GP|559557|gb|AAA51649.1|| arabinogalactan-pro... 31 2.0
AL368080 similar to GP|20259567|gb unknown protein {Arabidopsis ... 30 3.4
CA917688 weakly similar to GP|9294325|dbj| gene_id:K24M9.13~unkn... 30 4.5
BE942193 30 4.5
TC90660 similar to SP|O22176|WR15_ARATH Probable WRKY transcript... 30 4.5
TC86389 similar to GP|12322855|gb|AAG51417.1 unknown protein; 78... 29 5.9
TC87439 weakly similar to GP|5139695|dbj|BAA81686.1 expressed in... 29 5.9
TC80036 similar to GP|4512677|gb|AAD21731.1| unknown protein {Ar... 29 7.7
TC77505 MtN29 29 7.7
>BE248263 similar to GP|17528976|gb| unknown protein {Arabidopsis thaliana},
partial (9%)
Length = 306
Score = 135 bits (339), Expect = 8e-32
Identities = 66/66 (100%), Positives = 66/66 (100%)
Frame = +1
Query: 501 QRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQEWLVKNTKKVEKSIM 560
QRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQEWLVKNTKKVEKSIM
Sbjct: 1 QRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQEWLVKNTKKVEKSIM 180
Query: 561 LNGTTF 566
LNGTTF
Sbjct: 181 LNGTTF 198
Score = 79.0 bits (193), Expect = 6e-15
Identities = 40/54 (74%), Positives = 42/54 (77%)
Frame = +2
Query: 548 LVKNTKKVEKSIMLNGTTFIDKGPVASISTTSSKVVYPDTSKMLIYDPKHNPGT 601
L K +K + L FIDKGPVASISTTSSKVVYPDTSKMLIYDPKHNPGT
Sbjct: 143 LSKIRRKWRSLLCLMEQPFIDKGPVASISTTSSKVVYPDTSKMLIYDPKHNPGT 304
>BE323427 similar to GP|17528976|gb| unknown protein {Arabidopsis thaliana},
partial (8%)
Length = 251
Score = 107 bits (268), Expect(2) = 7e-29
Identities = 54/60 (90%), Positives = 54/60 (90%)
Frame = +1
Query: 441 PVYILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVE 500
P L DFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVE
Sbjct: 70 PFIFLNMPDFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVE 249
Score = 38.5 bits (88), Expect(2) = 7e-29
Identities = 16/16 (100%), Positives = 16/16 (100%)
Frame = +2
Query: 421 KDPKMAHNVFEAGLKH 436
KDPKMAHNVFEAGLKH
Sbjct: 8 KDPKMAHNVFEAGLKH 55
Score = 32.3 bits (72), Expect = 0.69
Identities = 14/15 (93%), Positives = 14/15 (93%)
Frame = +3
Query: 437 FMHEPVYILEYADFL 451
FMHEPVYILEYA FL
Sbjct: 57 FMHEPVYILEYA*FL 101
>TC91329 similar to GP|7682783|gb|AAF67364.1| Hypothetical protein T32B20.g
{Arabidopsis thaliana}, partial (38%)
Length = 1068
Score = 45.8 bits (107), Expect = 6e-05
Identities = 56/251 (22%), Positives = 100/251 (39%), Gaps = 28/251 (11%)
Frame = +3
Query: 284 SNKRVIFTYEQCLMY--LYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRY 341
+N RVIF + Y + + VW ++A K + A+ + +R+ A P E+ R
Sbjct: 75 ANARVIFDKAVQVNYKTVDNLASVWCEWAEIELKHENFKGALDLMRRAT-AEPSVEVKRK 251
Query: 342 A---------------------YAELEESRGAIQAAKKIYENLLGDSENATALAHIQFIR 380
+ +LEES G++++ +++YE +L D AT I +
Sbjct: 252 VAADGNQPVQMKLHKSLRLWTFFVDLEESLGSLESTREVYERIL-DLRIATPQIIINYAY 428
Query: 381 FLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMH- 439
FL KYF DA K V+E G+K F +
Sbjct: 429 FLEE-------HKYFEDAFK----------------------------VYERGVKIFKYP 503
Query: 440 --EPVYILEYADFLIRLNDD--QNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLAS 495
+ +++ + F+ R + R LFE A+ + P + ++ ++ K E+ YG
Sbjct: 504 HVKDIWVTYLSKFVKRYGRTKLERARELFENAVETAPADQVKPLYLQYAKLEEDYGLAKR 683
Query: 496 MLKVEQRRKEA 506
+KV + +A
Sbjct: 684 AMKVYDQATKA 716
Score = 41.2 bits (95), Expect = 0.001
Identities = 42/217 (19%), Positives = 86/217 (39%), Gaps = 8/217 (3%)
Frame = +3
Query: 301 HYPDVWYDYATWHAKA---GSIDAAIKVFQRSLKALPDSEM--LRYAYAELEESRGAIQA 355
H D+W Y + K ++ A ++F+ +++ P ++ L YA+LEE G +
Sbjct: 504 HVKDIWVTYLSKFVKRYGRTKLERARELFENAVETAPADQVKPLYLQYAKLEEDYGLAKR 683
Query: 356 AKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASV 415
A K+Y+ + T+ V K + Y +Y+A A+
Sbjct: 684 AMKVYD---------------------QATKAVPNNEKLSM---------YEIYIARAAE 773
Query: 416 AFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSSLPLED 475
F + K ++ E+GL L+YA+ L + + R ++ A
Sbjct: 774 IFGVPKTREIYEQAIESGLPD-KDVKTMCLKYAELERSLGEIERARGVYVFASKFADPRS 950
Query: 476 SVEVWKRFVKFEQTYGD---LASMLKVEQRRKEAFGE 509
+ W + +FE +G+ ML++++ ++ +
Sbjct: 951 DPDFWNDWHEFEVQHGNEDTFREMLRIKRSVSASYSQ 1061
>TC89278 similar to GP|9757719|dbj|BAB08244.1 CRN (crooked neck) protein
{Arabidopsis thaliana}, partial (64%)
Length = 1612
Score = 41.6 bits (96), Expect = 0.001
Identities = 43/167 (25%), Positives = 74/167 (43%), Gaps = 20/167 (11%)
Frame = +1
Query: 338 MLRYAYAELEESRGAIQAAKKIYENL---LGDSENATALAHIQFIRFLRRTEGVEPAR-- 392
+L Y YA+ E G + A+ +YE L D E A L + F F R + E AR
Sbjct: 28 VLGYRYAKFEMKNGEVPKARNVYERAVEKLADDEEA-ELLFVAFAEFEERCKEAERARCI 204
Query: 393 -KYFLD--ARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEA--GLKHFMHE------P 441
K+ LD + Y +VA+ +K + +A G + F +E P
Sbjct: 205 YKFALDHIPKGRAEDLYRKFVAF-------EKQYGDREGIEDAIVGKRRFQYEDEVRKNP 363
Query: 442 VYILEYADFLIRLNDD----QNIRALFERALSSLPLEDSVEVWKRFV 484
+ + D+ IRL + + R ++ERA++++P + W+R++
Sbjct: 364 LNYDSWFDY-IRLEESVGNKERTREVYERAIANVPPAEEKRYWQRYI 501
Score = 39.7 bits (91), Expect = 0.004
Identities = 44/198 (22%), Positives = 85/198 (42%), Gaps = 14/198 (7%)
Frame = +1
Query: 304 DVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRY---------AYAELEE-SRGAI 353
D W+DY G+ + +V++R++ +P +E RY YA EE G +
Sbjct: 373 DSWFDYIRLEESVGNKERTREVYERAIANVPPAEEKRYWQRYIYLWINYALYEELDAGDM 552
Query: 354 QAAKKIYENLLGDSENAT-ALAHIQFI--RFLRRTEGVEPARKYFLDA-RKSPSCTYHVY 409
+ + +Y+ L + + A I + +F R + AR+ +A K+P ++
Sbjct: 553 ERTRDVYKECLNQIPHQKFSFAKIWLLAAQFEIRQLNLTGARQILGNAIGKAPKDK--IF 726
Query: 410 VAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALS 469
Y + L + ++E L+ +YA+ L + + RA+FE A++
Sbjct: 727 KKYIEIELQLGNIDR-CRKLYEKYLEWSPENCYAWSKYAELERSLAETERARAIFELAIA 903
Query: 470 SLPLEDSVEVWKRFVKFE 487
L+ +WK ++ FE
Sbjct: 904 QPALDMPELMWKAYIDFE 957
>TC92163 similar to PIR|C84861|C84861 hypothetical protein At2g43040
[imported] - Arabidopsis thaliana, partial (48%)
Length = 1006
Score = 32.0 bits (71), Expect = 0.91
Identities = 57/238 (23%), Positives = 92/238 (37%), Gaps = 10/238 (4%)
Frame = +1
Query: 279 IDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEM 338
+ + S + +E + +YH D W A H AG +A+ + ++SL
Sbjct: 316 LSVCSQTSVLAMQFEDLMPGVYHRIDRWNSLALCHCAAGQNVSALNLLRKSL-------- 471
Query: 339 LRYAYAELEESRGAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDA 398
+ + ++ + AAK EN T LA EGV A++ +A
Sbjct: 472 --HKHERPDDLTSLLLAAKICSEN--------TCLA----------GEGVGHAQRAIKNA 591
Query: 399 RKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQ 458
P+ + VA + CL K K+A + FE F + + LE L + N D
Sbjct: 592 H-GPN-EHLKGVALRMLGLCLGKQAKVASSDFER--SRFQSKALESLEEGTRLEKNNSD- 756
Query: 459 NIRALFERALSSLP---LEDSVEVWKRFVKFEQTYGD-------LASMLKVEQRRKEA 506
+FE A+ L ++ + F F +T G LA +L +QR EA
Sbjct: 757 ---LIFELAVQYAEHRNLTSALRSARHF--FNETGGSVVKAWILLALILSAQQRFPEA 915
>TC77705 similar to GP|21553857|gb|AAM62950.1 unknown {Arabidopsis
thaliana}, partial (58%)
Length = 1139
Score = 31.2 bits (69), Expect = 1.5
Identities = 26/93 (27%), Positives = 45/93 (47%), Gaps = 2/93 (2%)
Frame = +1
Query: 624 LANLPSVDGPTPNVDIVLSICLQSDLPTGQSVKVGIPSQLPAGPAPATSELSGSSKSHPV 683
+ PSV T ++ S+ +SD+PT + + S+ APA +ELS S ++ V
Sbjct: 700 ITQAPSVSTET---ELSSSVPTESDVPTQTELSSSVQSE---SIAPAKTELSSSIQTESV 861
Query: 684 QSGLSHMQPGRKQYGK--RKQLDSQEEDDTKSV 714
+ S P + + Q++SQ + +T SV
Sbjct: 862 ATDASISVPPESSPAEVSQVQVESQIQTETSSV 960
>TC77143 similar to GP|559557|gb|AAA51649.1|| arabinogalactan-protein {Pyrus
communis}, partial (30%)
Length = 956
Score = 30.8 bits (68), Expect = 2.0
Identities = 36/129 (27%), Positives = 44/129 (33%), Gaps = 4/129 (3%)
Frame = +3
Query: 561 LNGTTFIDKGPVASISTTSSKVVYPDTSKMLIYDPKHNPGTGAAGTNAFDEILKATPPAL 620
L T+ + P A+ + S P T + P P T T ATPP
Sbjct: 150 LLATSCFAQAPGAAPTQPPSATPTPPTPAPVATPPTATPPTATPPT--------ATPPPA 305
Query: 621 VAFLANLP----SVDGPTPNVDIVLSICLQSDLPTGQSVKVGIPSQLPAGPAPATSELSG 676
A P S PTP SD PT S P+ P GPAP G
Sbjct: 306 AAPTPATPAPATSPPAPTPT----------SDAPTPDSTSSSPPAPGPGGPAPG----PG 443
Query: 677 SSKSHPVQS 685
S+ + P S
Sbjct: 444 STDAPPSPS 470
>AL368080 similar to GP|20259567|gb unknown protein {Arabidopsis thaliana},
partial (15%)
Length = 389
Score = 30.0 bits (66), Expect = 3.4
Identities = 12/43 (27%), Positives = 23/43 (52%)
Frame = +3
Query: 287 RVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRS 329
+++ YE+C++ +YP+ W Y + S+D A V R+
Sbjct: 249 KIVKLYERCVIACANYPEYWIRYVLCMEASESMDLANNVLARA 377
>CA917688 weakly similar to GP|9294325|dbj| gene_id:K24M9.13~unknown protein
{Arabidopsis thaliana}, partial (5%)
Length = 873
Score = 29.6 bits (65), Expect = 4.5
Identities = 19/60 (31%), Positives = 29/60 (47%), Gaps = 1/60 (1%)
Frame = +1
Query: 650 PTGQSVKVGIPSQLPA-GPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEE 708
P+ +V I SQL G + + G+S H L+H+QPG+ G + D+ EE
Sbjct: 694 PSKTIAEVEILSQLSTPGKRNFGNSIKGASSEHVKSDNLNHLQPGQNTVGHK---DNNEE 864
>BE942193
Length = 390
Score = 29.6 bits (65), Expect = 4.5
Identities = 15/35 (42%), Positives = 20/35 (56%)
Frame = +2
Query: 225 FLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFL 259
FLFL +YW+ L + LF N N+ P L LF+
Sbjct: 191 FLFLVQYWIPLTFLV---LFTNSNNVNPSRLVLFI 286
>TC90660 similar to SP|O22176|WR15_ARATH Probable WRKY transcription factor
15 (WRKY DNA-binding protein 15). [Mouse-ear cress],
partial (9%)
Length = 453
Score = 29.6 bits (65), Expect = 4.5
Identities = 15/35 (42%), Positives = 20/35 (56%)
Frame = +2
Query: 225 FLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFL 259
FLFL +YW+ L + LF N N+ P L LF+
Sbjct: 254 FLFLVQYWIPLTFLV---LFTNSNNVNPSRLVLFI 349
>TC86389 similar to GP|12322855|gb|AAG51417.1 unknown protein; 78656-75813
{Arabidopsis thaliana}, partial (86%)
Length = 1260
Score = 29.3 bits (64), Expect = 5.9
Identities = 37/165 (22%), Positives = 65/165 (38%), Gaps = 10/165 (6%)
Frame = +1
Query: 319 IDAAIKVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDSENATALAHIQF 378
+D A + K +S+ + A L E++G+ + A+K Y +LL D+ + H +
Sbjct: 355 LDVAQDCTKALRKRFSESKRVGRLEAMLLEAKGSWEKAEKAYSSLLEDNP-LDQIIHKRR 531
Query: 379 IRFLRRTEGVEPA----RKYFL------DARKSPSCTYHVYVAYASVAFCLDKDPKMAHN 428
+ + + A KY DA + + Y Y AFC ++
Sbjct: 532 VAMAKAQGNISGAIEWLNKYLEIFMADHDAWRELAEIYSSLQMYKQAAFCYEEVILAQPT 711
Query: 429 VFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSSLPL 473
V P+Y L YAD L L +N++ + S++ L
Sbjct: 712 V-----------PLYHLAYADVLYTLGGLENVQTAKKYYASTIDL 813
>TC87439 weakly similar to GP|5139695|dbj|BAA81686.1 expressed in cucumber
hypocotyls {Cucumis sativus}, partial (57%)
Length = 897
Score = 29.3 bits (64), Expect = 5.9
Identities = 33/110 (30%), Positives = 47/110 (42%), Gaps = 1/110 (0%)
Frame = +3
Query: 573 ASISTTSSKVVYPDTSKMLIYDPKHNPGTGAAGTNAFDEILKATPPALVAFLANLPSVDG 632
A+ + SS V P T+ P +P + T+ ATPPA + A P V
Sbjct: 144 AATTPVSSPVAAPPTTPTTPA-PVASPKSSPPATSPKAAAPTATPPAASSPPAVTP-VST 317
Query: 633 PTPNVDIVLSICLQSDLPTGQSVK-VGIPSQLPAGPAPATSELSGSSKSH 681
P P V S + + + +V V P+ PA PAPA S+ + K H
Sbjct: 318 PPPAPVPVKSPPTPAPVSSPPAVTPVAAPTTTPAVPAPAPSKGKKNKKKH 467
>TC80036 similar to GP|4512677|gb|AAD21731.1| unknown protein {Arabidopsis
thaliana}, partial (28%)
Length = 824
Score = 28.9 bits (63), Expect = 7.7
Identities = 14/49 (28%), Positives = 29/49 (58%), Gaps = 2/49 (4%)
Frame = +3
Query: 685 SGLSHMQPGRKQYGKRKQLDSQEEDDT--KSVQSQPLPQDAFRIRQFQK 731
S L + PG ++ GK+ + + +EE+D +SV +P +A+ + ++K
Sbjct: 510 SSLVEIIPGLQRLGKKNEEEEEEEEDVYDESVVQRPYLSEAWEVYDWRK 656
>TC77505 MtN29
Length = 889
Score = 28.9 bits (63), Expect = 7.7
Identities = 14/35 (40%), Positives = 21/35 (60%)
Frame = -1
Query: 564 TTFIDKGPVASISTTSSKVVYPDTSKMLIYDPKHN 598
T F+ P+ASIS +SS + +S+ IY P +N
Sbjct: 304 TIFVSSSPIASISPSSSFSTF-SSSQQFIYHPMYN 203
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,780,413
Number of Sequences: 36976
Number of extensions: 340440
Number of successful extensions: 1845
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 1825
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1843
length of query: 757
length of database: 9,014,727
effective HSP length: 103
effective length of query: 654
effective length of database: 5,206,199
effective search space: 3404854146
effective search space used: 3404854146
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146910.2


