

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146866.3 + phase: 0
(482 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
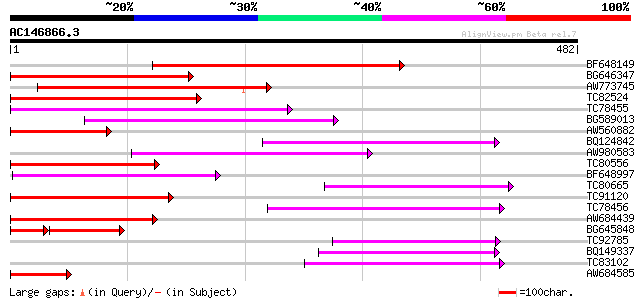
Score E
Sequences producing significant alignments: (bits) Value
BF648149 similar to PIR|T05135|T05 hypothetical protein F7H19.22... 278 3e-75
BG646347 similar to PIR|T05135|T05 hypothetical protein F7H19.22... 214 6e-56
AW773745 weakly similar to GP|19310633|gb unknown protein {Arabi... 153 1e-37
TC82524 similar to GP|19310633|gb|AAL85047.1 unknown protein {Ar... 144 6e-35
TC78455 similar to GP|11994404|dbj|BAB02363. gb|AAC28507.1~gene_... 141 5e-34
BG589013 weakly similar to GP|20805025|db contains EST D15170(C0... 139 3e-33
AW560882 similar to PIR|T05135|T05 hypothetical protein F7H19.22... 139 3e-33
BQ124842 weakly similar to GP|19310633|gb unknown protein {Arabi... 134 6e-32
AW980583 weakly similar to GP|16797803|db hypothetical membrane ... 123 2e-28
TC80556 weakly similar to PIR|F86285|F86285 hypothetical protein... 119 3e-27
BF648997 weakly similar to GP|13384114|gb aberrant lateral root ... 113 2e-25
TC80665 weakly similar to GP|19310633|gb|AAL85047.1 unknown prot... 112 4e-25
TC91120 similar to GP|11994404|dbj|BAB02363. gb|AAC28507.1~gene_... 101 6e-22
TC78456 similar to GP|11994404|dbj|BAB02363. gb|AAC28507.1~gene_... 99 4e-21
AW684439 weakly similar to PIR|F86285|F86 hypothetical protein A... 94 9e-20
BG645848 similar to GP|19310633|gb unknown protein {Arabidopsis ... 72 1e-19
TC92785 weakly similar to GP|13937234|gb|AAK50109.1 AT5g38030/F1... 91 1e-18
BQ149337 weakly similar to GP|13272459|gb unknown protein {Arabi... 89 5e-18
TC83102 similar to GP|11994404|dbj|BAB02363. gb|AAC28507.1~gene_... 87 1e-17
AW684585 similar to PIR|T05135|T051 hypothetical protein F7H19.2... 87 1e-17
>BF648149 similar to PIR|T05135|T05 hypothetical protein F7H19.220 -
Arabidopsis thaliana, partial (42%)
Length = 642
Score = 278 bits (711), Expect = 3e-75
Identities = 127/214 (59%), Positives = 166/214 (77%)
Frame = +1
Query: 122 LRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLYF 181
LR+Q ITLPLTY + +S+LLHIP+N+ LV Q+GI G+++ V TN NLV+ L +++
Sbjct: 1 LRSQSITLPLTYSATLSILLHIPINYFLVNVLQLGIRGIALGSVWTNFNLVVSLIIYIWV 180
Query: 182 SSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIASM 241
S +KK+W S C KGW SLL+LAIP+C+SVCLEWWWYE MI++CGLL+NP AT+ASM
Sbjct: 181 SGTHKKTWSGISSACFKGWKSLLNLAIPSCISVCLEWWWYEIMILLCGLLLNPHATVASM 360
Query: 242 GILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFTT 301
G+LIQTT+L+Y+FPSSLS GVSTR+GNELGA PQKA+++ IV L + VLG A+ F
Sbjct: 361 GVLIQTTALIYIFPSSLSFGVSTRVGNELGAENPQKAKLAAIVGLCFSFVLGFSALFFAF 540
Query: 302 LMRNQWGKFFTNDREILELTSIVLPIVGLCELGN 335
+RN W FT+D +I+ LTS+VLPI+GLC LGN
Sbjct: 541 SVRNIWATMFTSDPQIIALTSMVLPIIGLCXLGN 642
>BG646347 similar to PIR|T05135|T05 hypothetical protein F7H19.220 -
Arabidopsis thaliana, partial (33%)
Length = 752
Score = 214 bits (545), Expect = 6e-56
Identities = 102/156 (65%), Positives = 131/156 (83%)
Frame = +3
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
MISM+FLG+L ++ LAGGSL+IGFANITGYS++SGLA+GMEPICGQA+GAK++ +LGL L
Sbjct: 279 MISMLFLGHLNDLALAGGSLAIGFANITGYSILSGLAVGMEPICGQAFGAKKFTLLGLCL 458
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
QRT+LLLL SIPISF W+ MK++LLF QD EI++MAQ++IL+ +PDL S +HPLRI
Sbjct: 459 QRTILLLLLVSIPISFSWLYMKKMLLFFNQDEEIATMAQTYILYSIPDLIAQSFIHPLRI 638
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMG 156
YLRTQ ITLPLT C+ S+LLHIP+N+ L + +G
Sbjct: 639 YLRTQSITLPLTLCATFSILLHIPINYFLGIISYLG 746
>AW773745 weakly similar to GP|19310633|gb unknown protein {Arabidopsis
thaliana}, partial (36%)
Length = 614
Score = 153 bits (387), Expect = 1e-37
Identities = 79/201 (39%), Positives = 125/201 (61%), Gaps = 2/201 (0%)
Frame = +3
Query: 24 FANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTLQRTVLLLLSTSIPISFIWINMKR 83
FA++TG+S++ G+A ++ +CGQ+YGAKQ+++LG+ +QR + +L+ +IP++ IW N +
Sbjct: 12 FASVTGFSLLVGMASALDTLCGQSYGAKQYRMLGIHMQRAMFILMIVAIPLAIIWANTRS 191
Query: 84 ILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRIYLRTQGITLPLTYCSAVSVLLHI 143
IL+F GQD EIS A ++ +VP LF +L L +L+TQ I P+ SAV+ LLH+
Sbjct: 192 ILIFLGQDHEISMEAGNYAKLMVPSLFAYGLLQCLNRFLQTQNIVFPMMLSSAVTTLLHL 371
Query: 144 PLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLYFSSVYKKSWISPSLDCI--KGWS 201
PL + +V G G +IA ++ VI+LS ++ FS +K+W S + +
Sbjct: 372 PLCWYMVYKSGFGSGGAAIASSISYWVNVIILSLYVKFSPSCQKTWNGFSREALAPNNIP 551
Query: 202 SLLSLAIPTCVSVCLEWWWYE 222
L LAIP+ VCLE W +E
Sbjct: 552 IFLKLAIPSAAMVCLEIWSFE 614
>TC82524 similar to GP|19310633|gb|AAL85047.1 unknown protein {Arabidopsis
thaliana}, partial (36%)
Length = 712
Score = 144 bits (364), Expect = 6e-35
Identities = 67/163 (41%), Positives = 112/163 (68%)
Frame = +2
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
+IS++F+G+LGE+ L+G S++ FA++TG+S++ G+A ++ CGQ+YGAKQ+++LG+ +
Sbjct: 206 LISVMFVGHLGELPLSGASMATSFASVTGFSLLQGMASALDTFCGQSYGAKQYRMLGVHM 385
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
QR + +L+ +IP++ IW N + ILL GQD EIS A S+ +VP LF +L L
Sbjct: 386 QRAMFILMVVAIPLAVIWANTRSILLVLGQDPEISIEAGSYAKLMVPCLFAYGLLQCLNR 565
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIA 163
+L+TQ I P+ + SA++ LLH+P+ + +V +G +IA
Sbjct: 566 FLQTQNIVFPMMFSSAMTTLLHLPICWFMVYKSGLGSRXAAIA 694
>TC78455 similar to GP|11994404|dbj|BAB02363.
gb|AAC28507.1~gene_id:MIL23.25~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (48%)
Length = 1060
Score = 141 bits (356), Expect = 5e-34
Identities = 74/241 (30%), Positives = 134/241 (54%), Gaps = 1/241 (0%)
Frame = +1
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
M + IF G+LG +ELA SL I Y ++ G+ +E +CGQA+GAK++++LG+ L
Sbjct: 277 MSTQIFSGHLGNLELAAASLGNNGIQIFAYGLMLGMGSAVETLCGQAFGAKKYEMLGIYL 456
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
QR+ +LL + ++ I+I + IL+F G+ +I+S A F+ L+P +F +I P++
Sbjct: 457 QRSTVLLTIAGLILTIIYIFSEPILIFLGESPKIASAASLFVFGLIPQIFAYAINFPIQK 636
Query: 121 YLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLY 180
+L+ Q I P Y SA ++++H+ L+++++ +G+ G S+ + ++ +VI ++
Sbjct: 637 FLQAQSIVAPSAYISAATLVIHLVLSYVVIYQIGLGLLGASLVLSISWWIIVIAQFVYIV 816
Query: 181 FSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWY-EFMIMMCGLLVNPKATIA 239
S K +W S G L+ + V +CLE W+ GLL +P+ +
Sbjct: 817 KSEKCKHTWKGFSFQAFSGLPEFFKLSAASAVMLCLETWYLSNLXFCXAGLLPHPELALD 996
Query: 240 S 240
S
Sbjct: 997 S 999
>BG589013 weakly similar to GP|20805025|db contains EST D15170(C0202)~unknown
protein {Oryza sativa (japonica cultivar-group)},
partial (42%)
Length = 786
Score = 139 bits (349), Expect = 3e-33
Identities = 71/216 (32%), Positives = 122/216 (55%)
Frame = +1
Query: 64 VLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRIYLR 123
+L L PIS +WI +IL+F QD +S +A+ + ++L+P LF ++L L Y +
Sbjct: 34 ILTLTVVCFPISLVWIFTDKILMFFSQDPGMSHVAREYCIYLIPALFGYALLQALIRYFQ 213
Query: 124 TQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFLYFSS 183
TQG+ P+ + S ++ LHIP+ ++LV +G G ++A+ ++ VI L ++ +S
Sbjct: 214 TQGMIFPMVFSSVSALFLHIPICWILVFKLGLGHIGAALAIGISYWLNVIWLWVYIKYSP 393
Query: 184 VYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEFMIMMCGLLVNPKATIASMGI 243
+K+ I S + AIP+ + C EWW +E +I++ GLL NP+ + + +
Sbjct: 394 SCEKTKIVFSTHALHNLPEFCKYAIPSGLMFCFEWWSFEILILIAGLLPNPQLETSVLSV 573
Query: 244 LIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKAR 279
+ TTSL + P ++ STR+ NELGA P+ A+
Sbjct: 574 CLNTTSLHFFIPYAIGASASTRVSNELGAGNPKTAK 681
>AW560882 similar to PIR|T05135|T05 hypothetical protein F7H19.220 -
Arabidopsis thaliana, partial (22%)
Length = 582
Score = 139 bits (349), Expect = 3e-33
Identities = 65/86 (75%), Positives = 81/86 (93%)
Frame = +2
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
+ISM+FLG +GE+ LAGGSL+IGFANITGYS++SGLAMGMEPICGQA+GAK++K+LGLT+
Sbjct: 323 IISMLFLGRVGELALAGGSLAIGFANITGYSILSGLAMGMEPICGQAFGAKRFKLLGLTM 502
Query: 61 QRTVLLLLSTSIPISFIWINMKRILL 86
QRT +LLL TSI ISF+W+NMKR+LL
Sbjct: 503 QRTAILLLVTSIVISFLWLNMKRLLL 580
>BQ124842 weakly similar to GP|19310633|gb unknown protein {Arabidopsis
thaliana}, partial (31%)
Length = 706
Score = 134 bits (338), Expect = 6e-32
Identities = 66/201 (32%), Positives = 114/201 (55%)
Frame = +2
Query: 216 LEWWWYEFMIMMCGLLVNPKATIASMGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRP 275
L+ W +E M++M GLL NP + + I + T L ++ P S VS R+ NELG P
Sbjct: 95 LKVWTFELMVLMSGLLPNPVIETSVLSICLNTFGLAWMIPFGCSCAVSIRVSNELGGGNP 274
Query: 276 QKARISMIVSLFLAMVLGLGAMLFTTLMRNQWGKFFTNDREILELTSIVLPIVGLCELGN 335
A +++ V+L ++ + L +L L R WG +++D++++ S ++PI+ + +
Sbjct: 275 NGASLAVRVALSISFIAALFMVLSMILARKVWGHLYSDDKQVIRYVSAMMPILAISSFLD 454
Query: 336 CPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSC 395
Q+T GVL G IGA +NLGSFY+VG+P A+ L F + GLW+G+++A
Sbjct: 455 AIQSTLSGVLAGCGWQKIGAYVNLGSFYVVGVPCAVVLAFFVHMHAMGLWLGIISAFIVQ 634
Query: 396 AMLMLVVLCRTDWNLQVQRAK 416
L ++ R++W + ++A+
Sbjct: 635 TSLYIIFTIRSNWEEEAKKAQ 697
>AW980583 weakly similar to GP|16797803|db hypothetical membrane protein-1
{Marchantia polymorpha}, partial (21%)
Length = 631
Score = 123 bits (308), Expect = 2e-28
Identities = 65/205 (31%), Positives = 108/205 (51%)
Frame = +3
Query: 104 FLVPDLFLLSILHPLRIYLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIA 163
+L+P++ IL + +L+TQ I PL S ++ LLH ++ +V + GI G +IA
Sbjct: 15 YLIPNILASGILKCIVKFLQTQNIVFPLLVASGITSLLHCLNCWIWIVKLRHGIKGAAIA 194
Query: 164 MVLTNLNLVILLSSFLYFSSVYKKSWISPSLDCIKGWSSLLSLAIPTCVSVCLEWWWYEF 223
++N +LL ++ FSS K +W S + + L +A P+ + VCLE W YE
Sbjct: 195 TCMSNWTYTVLLVFYIKFSSSCKSTWTGFSRESLHNIPQFLRIAFPSAIMVCLESWMYEI 374
Query: 224 MIMMCGLLVNPKATIASMGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKARISMI 283
M+++ G L NPK + + I + S+V++ S + S R+ NELGA P+ A +++
Sbjct: 375 MVLLSGTLPNPKLQTSVLAICMNIASVVWMLSSGFTGAASIRVSNELGAGNPRAAHLAVC 554
Query: 284 VSLFLAMVLGLGAMLFTTLMRNQWG 308
V + L + L+RN WG
Sbjct: 555 VVVVLNITEAFFVGTVMILLRNIWG 629
>TC80556 weakly similar to PIR|F86285|F86285 hypothetical protein AAD39644.1
[imported] - Arabidopsis thaliana, partial (31%)
Length = 661
Score = 119 bits (297), Expect = 3e-27
Identities = 54/127 (42%), Positives = 89/127 (69%)
Frame = +1
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
++S++ +G+LGE+ L+ +L+I FA +TG+S + G+A G+E CGQAYGAKQ++ +G+
Sbjct: 277 VVSIMIVGHLGELYLSSAALAISFAGVTGFSFLMGMASGLETTCGQAYGAKQYQRIGIQT 456
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
++ L+ +P+SFIWIN++ IL+F+GQD I+ A F ++L+P LF +IL PL
Sbjct: 457 YTSIFSLILVCLPLSFIWINIESILVFTGQDPLIAHEAGRFTIWLIPALFAYAILQPLVR 636
Query: 121 YLRTQGI 127
Y + Q +
Sbjct: 637 YFQIQSL 657
>BF648997 weakly similar to GP|13384114|gb aberrant lateral root formation 5
{Arabidopsis thaliana}, partial (27%)
Length = 536
Score = 113 bits (282), Expect = 2e-25
Identities = 59/177 (33%), Positives = 106/177 (59%)
Frame = +3
Query: 3 SMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTLQR 62
S++ +G+LGE++LAG +L+ + ++TG +V+ GL+ +E +CGQ +GAK++ +LG+ LQ
Sbjct: 3 SVMLVGHLGELQLAGATLANSWFSVTGVAVMVGLSGALETLCGQGFGAKEYHMLGIYLQG 182
Query: 63 TVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRIYL 122
+ ++ SI IS IW + IL+ Q +I+ A ++ FL+P LF SIL L +L
Sbjct: 183 SCIISFIFSIFISIIWFYTEHILVLLHQSQDIARTAALYMKFLIPGLFAYSILQNLLRFL 362
Query: 123 RTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLVILLSSFL 179
+TQ + +PL SA+ L+H+ + + V + G +A + +I+L ++
Sbjct: 363 QTQSVVMPLVILSAIPTLIHVGIAYGFVQWTGLNFIGGPVATSIXLWISMIMLXFYV 533
>TC80665 weakly similar to GP|19310633|gb|AAL85047.1 unknown protein
{Arabidopsis thaliana}, partial (31%)
Length = 894
Score = 112 bits (279), Expect = 4e-25
Identities = 54/161 (33%), Positives = 95/161 (58%)
Frame = +3
Query: 268 NELGANRPQKARISMIVSLFLAMVLGLGAMLFTTLMRNQWGKFFTNDREILELTSIVLPI 327
NELGA P+ AR+++ V + +A++ + L+RN WG ++N+ E+++ +I+LPI
Sbjct: 3 NELGAGNPRAARLAVYVVVVIAIIESIVVGAVIILIRNIWGYAYSNEEEVVKYVAIMLPI 182
Query: 328 VGLCELGNCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLWIG 387
+ + + Q+ G RG IGA +NLGS+YLVG+P A+ L FV +G GLW+G
Sbjct: 183 IAVSNFLDGIQSVLSGTARGVGWQKIGAYVNLGSYYLVGIPAAVVLAFVLHVGGKGLWLG 362
Query: 388 LLAAQGSCAMLMLVVLCRTDWNLQVQRAKELTKSSTTSDDV 428
++ A + + ++ RTDW + ++A + S T++ +
Sbjct: 363 IICALFVQVVSLTIITIRTDWEKEAKKATDRVYDSITTESL 485
>TC91120 similar to GP|11994404|dbj|BAB02363.
gb|AAC28507.1~gene_id:MIL23.25~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (35%)
Length = 665
Score = 101 bits (252), Expect = 6e-22
Identities = 53/139 (38%), Positives = 84/139 (60%)
Frame = +3
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
M + IF G+LG +ELA SL I Y ++ G+ +E +CGQAYGA+++ +LG L
Sbjct: 249 MSTQIFSGHLGNLELAAASLGNTGIQIFAYGLMLGMGSAVETLCGQAYGAEKYGMLGTYL 428
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
QR+ +LL T ++ I++ + IL+F GQ I+S A F+ L+P +F ++ P++
Sbjct: 429 QRSTILLTITGFFLTIIYVLSEPILVFIGQSPRIASAAALFVYGLIPQIFAYAVNFPIQK 608
Query: 121 YLRTQGITLPLTYCSAVSV 139
+L+ Q I LP Y SA S+
Sbjct: 609 FLQAQSIVLPSAYISAGSL 665
>TC78456 similar to GP|11994404|dbj|BAB02363.
gb|AAC28507.1~gene_id:MIL23.25~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (40%)
Length = 920
Score = 99.0 bits (245), Expect = 4e-21
Identities = 55/202 (27%), Positives = 99/202 (48%), Gaps = 1/202 (0%)
Frame = +1
Query: 220 WYEFMIMMCGLLVNPKATIASMGILIQTTSLVYVFPSSLSLGVSTRIGNELGANRPQKAR 279
+++ ++++ GLL +P+ + S+ I + ++ S R+ NELGA + A
Sbjct: 1 YFQILVLLAGLLPHPELALDSLSICTTVSGWTFMISVGFQAAASVRVSNELGAGNSKSAS 180
Query: 280 ISMIVSLFLAMVLGLGAMLFTTLMRNQWGKFFTNDREILELTSIVLPIVGLCELGNCPQT 339
S++V ++ ++ L +R+ FT E+ S + P++ L + N Q
Sbjct: 181 FSVVVVTVISFIICAIIALVVLALRDVISYVFTEGEEVAAAVSNLSPLLALAIVLNGVQP 360
Query: 340 TGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGSCAMLM 399
GV G T A +N+G +Y +G+P+ LGF K G G+W+G+L +++
Sbjct: 361 VLSGVAVGCGWQTFVAYVNVGCYYGLGIPLGAVLGFYFKFGAKGIWLGMLGGTVLQTIIL 540
Query: 400 LVVLCRTDWNLQ-VQRAKELTK 420
+ V RTDWN + V+ K L K
Sbjct: 541 MWVTFRTDWNNEVVESNKRLNK 606
>AW684439 weakly similar to PIR|F86285|F86 hypothetical protein AAD39644.1
[imported] - Arabidopsis thaliana, partial (32%)
Length = 510
Score = 94.4 bits (233), Expect = 9e-20
Identities = 46/125 (36%), Positives = 76/125 (60%)
Frame = +3
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQWKILGLTL 60
+ISMI +G LG++ L+ +++I ++G+S++ G++ +E CGQAYGAKQ+K G+ +
Sbjct: 132 IISMIMVGRLGKLALSSTAIAISLCGVSGFSLLFGMSCALETQCGQAYGAKQYKKFGVQI 311
Query: 61 QRTVLLLLSTSIPISFIWINMKRILLFSGQDLEISSMAQSFILFLVPDLFLLSILHPLRI 120
V L+ +P+S +WI + R+L+ GQD IS A F + ++P LF + L L
Sbjct: 312 YTAVFSLIIACLPLSLLWIFLGRLLILLGQDPLISQEAGKFSMCMIPALFAYATLXALVX 491
Query: 121 YLRTQ 125
Y Q
Sbjct: 492 YFLMQ 506
>BG645848 similar to GP|19310633|gb unknown protein {Arabidopsis thaliana},
partial (24%)
Length = 682
Score = 71.6 bits (174), Expect(2) = 1e-19
Identities = 32/63 (50%), Positives = 45/63 (70%)
Frame = +1
Query: 35 GLAMGMEPICGQAYGAKQWKILGLTLQRTVLLLLSTSIPISFIWINMKRILLFSGQDLEI 94
G+ +E +CGQAYGAKQ+ +LG+ QR +L+LL SIP+S IW N +L+ GQD EI
Sbjct: 460 GMGSALETLCGQAYGAKQYHMLGVHTQRAMLVLLVLSIPLSLIWFNTSNLLIALGQDYEI 639
Query: 95 SSM 97
S++
Sbjct: 640 STL 648
Score = 43.1 bits (100), Expect(2) = 1e-19
Identities = 18/33 (54%), Positives = 28/33 (84%)
Frame = +2
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVI 33
MIS++F+G+LG++ L+G SL FA++TGYSV+
Sbjct: 362 MISIMFVGHLGKLPLSGASLGNSFASVTGYSVL 460
Score = 40.8 bits (94), Expect = 0.001
Identities = 19/77 (24%), Positives = 43/77 (55%)
Frame = +1
Query: 113 SILHPLRIYLRTQGITLPLTYCSAVSVLLHIPLNFLLVVHFQMGIAGVSIAMVLTNLNLV 172
+I+ L +L+TQ LP+ S ++ L+H+ ++ V +++GI G ++A+ L+ V
Sbjct: 7 AIIQCLNRFLQTQNNVLPMLISSGITTLVHLVFCWVFVFEYELGIKGAALAISLSYWVNV 186
Query: 173 ILLSSFLYFSSVYKKSW 189
+L ++ ++ +W
Sbjct: 187 FMLVIYINSATACASTW 237
>TC92785 weakly similar to GP|13937234|gb|AAK50109.1 AT5g38030/F16F17_30
{Arabidopsis thaliana}, partial (30%)
Length = 533
Score = 90.5 bits (223), Expect = 1e-18
Identities = 41/143 (28%), Positives = 83/143 (57%)
Frame = +1
Query: 275 PQKARISMIVSLFLAMVLGLGAMLFTTLMRNQWGKFFTNDREILELTSIVLPIVGLCELG 334
P+ AR S++V++ ++++GL L + R+++ +FT D+E+ +L + P++ LC +
Sbjct: 1 PRTARFSLVVAVITSILIGLLLALVLIISRDKYPAYFTTDKEVQDLVKDLTPLLALCVVI 180
Query: 335 NCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFPGLWIGLLAAQGS 394
N Q GV G+ A +N+ +YL G+PV + LG+ LG G+W G+++
Sbjct: 181 NNVQPVLSGVAIGAGWQAAVAYVNIACYYLFGIPVGLILGYKVNLGVKGIWCGMMSGTIL 360
Query: 395 CAMLMLVVLCRTDWNLQVQRAKE 417
++L+++ +T+WN + A++
Sbjct: 361 QTCVLLIMVYKTNWNKEASLAED 429
>BQ149337 weakly similar to GP|13272459|gb unknown protein {Arabidopsis
thaliana}, partial (21%)
Length = 622
Score = 88.6 bits (218), Expect = 5e-18
Identities = 44/154 (28%), Positives = 82/154 (52%)
Frame = +2
Query: 263 STRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFTTLMRNQWGKFFTNDREILELTS 322
S R+ NELG + A+ S++++ + +G L ++ + FT+D ++
Sbjct: 17 SVRVANELGRGSSRDAKFSIVINALTSFAIGFIFFLIFLFLKKKLSYIFTSDSDVANAVG 196
Query: 323 IVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVAIFLGFVAKLGFP 382
+ + L L N Q GV G+ +I A +N+G +YL+G+PV + +G V LG
Sbjct: 197 DLSFWLALSMLLNSVQPVLSGVSVGAGWQSIVAYVNIGCYYLIGIPVGVVIGVVYNLGIK 376
Query: 383 GLWIGLLAAQGSCAMLMLVVLCRTDWNLQVQRAK 416
G+WIG+L ++++++ +TDW+ QV+ A+
Sbjct: 377 GIWIGMLFGTFVQTIMLIIITTKTDWDKQVEIAQ 478
>TC83102 similar to GP|11994404|dbj|BAB02363.
gb|AAC28507.1~gene_id:MIL23.25~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (36%)
Length = 1048
Score = 87.4 bits (215), Expect = 1e-17
Identities = 50/171 (29%), Positives = 83/171 (48%), Gaps = 1/171 (0%)
Frame = +3
Query: 251 VYVFPSSLSLGVSTRIGNELGANRPQKARISMIVSLFLAMVLGLGAMLFTTLMRNQWGKF 310
V++ + S R+ NELGA P+ A S+ V ++ ++ + A L +R+
Sbjct: 231 VFMISVGFNAAASVRVSNELGARNPKSASFSVKVVTVISFIISVIAALIVLALRDVISYV 410
Query: 311 FTNDREILELTSIVLPIVGLCELGNCPQTTGCGVLRGSARPTIGANINLGSFYLVGMPVA 370
FT + S + P++ L + N Q GV G A +N+G +Y+ G+P+
Sbjct: 411 FTEGEVVAAAVSDLCPLLSLSLVLNGIQPVLSGVAVGCGWQAFVAYVNVGCYYIAGIPLG 590
Query: 371 IFLGFVAKLGFPGLWIGLLAAQGSCAMLMLVVLCRTDWNLQV-QRAKELTK 420
LGF G G+W+G+L ++++ V RTDWN +V + AK L K
Sbjct: 591 AGLGFYFNFGAKGIWLGMLGGTTMQTIILMWVTFRTDWNKEVKEAAKRLNK 743
>AW684585 similar to PIR|T05135|T051 hypothetical protein F7H19.220 -
Arabidopsis thaliana, partial (15%)
Length = 458
Score = 87.4 bits (215), Expect = 1e-17
Identities = 40/52 (76%), Positives = 49/52 (93%)
Frame = +3
Query: 1 MISMIFLGYLGEMELAGGSLSIGFANITGYSVISGLAMGMEPICGQAYGAKQ 52
MISM+FLG+LGE+ LAGGSL++GFANITGYS++SGLA GMEPI GQA+GAK+
Sbjct: 303 MISMLFLGHLGELALAGGSLAVGFANITGYSILSGLAXGMEPIXGQAFGAKR 458
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.141 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,672,482
Number of Sequences: 36976
Number of extensions: 273591
Number of successful extensions: 2276
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 2235
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2270
length of query: 482
length of database: 9,014,727
effective HSP length: 100
effective length of query: 382
effective length of database: 5,317,127
effective search space: 2031142514
effective search space used: 2031142514
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC146866.3


