

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146864.1 + phase: 0
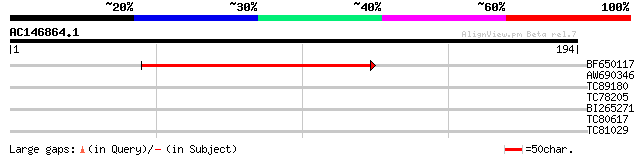
(194 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF650117 similar to PIR|C86187|C861 YUP8H12.12 [imported] - Arab... 97 5e-21
AW690346 weakly similar to GP|10177738|dbj gene_id:MSG15.11~pir|... 34 0.041
TC89180 weakly similar to GP|13174241|gb|AAK14415.1 putative glu... 29 1.0
TC78205 homologue to PIR|T47559|T47559 60S ribosomal protein-lik... 29 1.3
BI265271 weakly similar to PIR|F86151|F861 hypothetical protein ... 27 5.0
TC80617 weakly similar to PIR|F96672|F96672 Similar to Flavonol ... 27 6.5
TC81029 similar to GP|21553677|gb|AAM62770.1 unknown {Arabidopsi... 26 8.5
>BF650117 similar to PIR|C86187|C861 YUP8H12.12 [imported] - Arabidopsis
thaliana, partial (25%)
Length = 604
Score = 96.7 bits (239), Expect = 5e-21
Identities = 46/80 (57%), Positives = 63/80 (78%)
Frame = +2
Query: 46 QRRLELPKELTKNVIQLSCDSMEKGGVCDVYLVGTFHGNKESSKQVEEIVKFLKPEIVFL 105
++++ LP+EL++NVI LSC+S +GGVCDVYLVGT H ++ESSK+V+ IV LKPE VFL
Sbjct: 311 RQKVTLPEELSRNVIVLSCESSAEGGVCDVYLVGTAHVSEESSKEVQAIVSLLKPEAVFL 490
Query: 106 ELCSDRQEVLLHDNMEVSTI 125
ELCS R +VL N++ T+
Sbjct: 491 ELCSSRVQVLTLQNLKXPTM 550
>AW690346 weakly similar to GP|10177738|dbj
gene_id:MSG15.11~pir||C69291~similar to unknown protein
{Arabidopsis thaliana}, partial (22%)
Length = 338
Score = 33.9 bits (76), Expect = 0.041
Identities = 14/29 (48%), Positives = 22/29 (75%)
Frame = +2
Query: 75 VYLVGTFHGNKESSKQVEEIVKFLKPEIV 103
++L+GT H +KES+ VE +VK +KP+ V
Sbjct: 251 IWLIGTTHVSKESAMDVERVVKAVKPDNV 337
>TC89180 weakly similar to GP|13174241|gb|AAK14415.1 putative glutamine
synthetase {Oryza sativa}, partial (26%)
Length = 989
Score = 29.3 bits (64), Expect = 1.0
Identities = 11/25 (44%), Positives = 16/25 (64%)
Frame = -3
Query: 14 LRKVIQTTFTTLPCFSTTSAAFCRD 38
LRK++ F +PCFS S FC++
Sbjct: 225 LRKLLINNFVKIPCFSFLSQHFCKE 151
>TC78205 homologue to PIR|T47559|T47559 60S ribosomal protein-like -
Arabidopsis thaliana, partial (93%)
Length = 858
Score = 28.9 bits (63), Expect = 1.3
Identities = 22/55 (40%), Positives = 28/55 (50%), Gaps = 6/55 (10%)
Frame = +2
Query: 9 CLSTRLRKVIQTT------FTTLPCFSTTSAAFCRDQSPEKERQRRLELPKELTK 57
CLSTR K I TT LPC + + R P+K +R +ELPK LT+
Sbjct: 149 CLSTRNVKGISTTS*SPQSLHGLPCIGSNT----RRILPKKL*KRGVELPKSLTQ 301
>BI265271 weakly similar to PIR|F86151|F861 hypothetical protein AAF76475.1
[imported] - Arabidopsis thaliana, partial (16%)
Length = 662
Score = 26.9 bits (58), Expect = 5.0
Identities = 15/43 (34%), Positives = 24/43 (54%), Gaps = 4/43 (9%)
Frame = +3
Query: 37 RDQSPEKERQRRLELPKELTKN----VIQLSCDSMEKGGVCDV 75
R+ E+++Q +LP + K + Q+ C S EKG +CDV
Sbjct: 450 RNDITEEQKQAIAKLPFRMEKRCKAVMRQIICFSEEKGRLCDV 578
>TC80617 weakly similar to PIR|F96672|F96672 Similar to Flavonol
3-O-Glucosyltransferase [imported] - Arabidopsis
thaliana, partial (32%)
Length = 941
Score = 26.6 bits (57), Expect = 6.5
Identities = 10/21 (47%), Positives = 17/21 (80%)
Frame = +1
Query: 86 ESSKQVEEIVKFLKPEIVFLE 106
++ KQ+E ++K LKP+IVF +
Sbjct: 286 QTDKQIELLLKELKPQIVFFD 348
>TC81029 similar to GP|21553677|gb|AAM62770.1 unknown {Arabidopsis
thaliana}, partial (43%)
Length = 783
Score = 26.2 bits (56), Expect = 8.5
Identities = 19/60 (31%), Positives = 33/60 (54%), Gaps = 2/60 (3%)
Frame = -2
Query: 43 KERQRRLELPKELTKNVIQLSCDSMEK--GGVCDVYLVGTFHGNKESSKQVEEIVKFLKP 100
K++Q++ ++ ++L K +IQL CD E G + + G + K+ K+V E K KP
Sbjct: 335 KQQQKKKKMKQQL*KQLIQLVCDCCEPE*TGSLNWHRTGLYPKAKQ-RKRVAEKRKKRKP 159
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,908,308
Number of Sequences: 36976
Number of extensions: 94099
Number of successful extensions: 715
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 713
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 715
length of query: 194
length of database: 9,014,727
effective HSP length: 91
effective length of query: 103
effective length of database: 5,649,911
effective search space: 581940833
effective search space used: 581940833
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146864.1


