

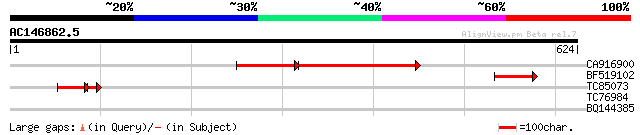
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146862.5 + phase: 0 /pseudo
(624 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CA916900 homologue to GP|6681366|db retinoblastoma-related prote... 270 e-107
BF519102 homologue to GP|6681366|db retinoblastoma-related prote... 94 2e-19
TC85073 similar to GP|6681366|dbj|BAA88690.1 retinoblastoma-rela... 60 3e-10
TC76984 similar to GP|13877717|gb|AAK43936.1 OsNAC6 protein-like... 32 0.95
BQ144385 similar to GP|21740500|emb OSJNBa0006B20.15 {Oryza sati... 29 6.2
>CA916900 homologue to GP|6681366|db retinoblastoma-related protein {Pisum
sativum}, partial (23%)
Length = 727
Score = 270 bits (689), Expect(2) = e-107
Identities = 133/135 (98%), Positives = 134/135 (98%)
Frame = +3
Query: 318 LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ 377
LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ
Sbjct: 321 LKSKLLPPPLQSAFASPTKPNPGGGGETCAETGISVFFSKIVKLGAVRISGMVERLQLSQ 500
Query: 378 QIRENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQC 437
QIRENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQ
Sbjct: 501 QIRENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKISQLNLTFREIIYNYRKQPQW 680
Query: 438 KPQVFRSVFVDWSSA 452
+PQVFRSVFVDWSSA
Sbjct: 681 QPQVFRSVFVDWSSA 725
Score = 139 bits (349), Expect(2) = e-107
Identities = 69/70 (98%), Positives = 69/70 (98%)
Frame = +2
Query: 250 FGELGMGKRFFDV*FISSR*TCPFCRNQSAWYVSRTNAIIR*NCNAN*LLLWRTASCAFL 309
FGELGMGKRFFDV*FISSR*TCPFCRNQSAWYVSRTNAIIR*NCNAN*LLLWRTASCAFL
Sbjct: 2 FGELGMGKRFFDV*FISSR*TCPFCRNQSAWYVSRTNAIIR*NCNAN*LLLWRTASCAFL 181
Query: 310 A*A*DFACLK 319
A*A*DFAC K
Sbjct: 182 A*A*DFACSK 211
>BF519102 homologue to GP|6681366|db retinoblastoma-related protein {Pisum
sativum}, partial (10%)
Length = 624
Score = 93.6 bits (231), Expect = 2e-19
Identities = 45/47 (95%), Positives = 45/47 (95%)
Frame = -1
Query: 534 VSAAHNVYVSPLRSSKMDALISHSSKSYYACXGESTHAYQSPXKDLT 580
VSAAHNVYVSPLRSSKMDALISHSSKSYYAC GESTHAYQSP KDLT
Sbjct: 624 VSAAHNVYVSPLRSSKMDALISHSSKSYYACVGESTHAYQSPSKDLT 484
>TC85073 similar to GP|6681366|dbj|BAA88690.1 retinoblastoma-related protein
{Pisum sativum}, partial (22%)
Length = 716
Score = 60.1 bits (144), Expect(2) = 3e-10
Identities = 31/35 (88%), Positives = 31/35 (88%)
Frame = +1
Query: 53 AFQVAQIQRLLPPPVSTAMTTAKWLRTVISPLPIK 87
AFQVAQIQRLLP VSTAMTTAKWLRTVISPL K
Sbjct: 571 AFQVAQIQRLLPXXVSTAMTTAKWLRTVISPLSSK 675
Score = 23.1 bits (48), Expect(2) = 3e-10
Identities = 11/16 (68%), Positives = 11/16 (68%)
Frame = +3
Query: 86 IKAVTRARAILDFMRQ 101
IK ARAILD MRQ
Sbjct: 669 IKXXXXARAILDXMRQ 716
>TC76984 similar to GP|13877717|gb|AAK43936.1 OsNAC6 protein-like protein
{Arabidopsis thaliana}, partial (71%)
Length = 1450
Score = 31.6 bits (70), Expect = 0.95
Identities = 28/84 (33%), Positives = 39/84 (46%), Gaps = 1/84 (1%)
Frame = -1
Query: 359 VKLGAVRISGMVERLQLSQQI-RENVYSLFQRILNQWTSLFFNRHIDQIILCCFYGVAKI 417
V GA R+ R+QL I +V + ++LN W L F II+ + V I
Sbjct: 793 VPTGAARV-----RVQLWN*I*SVHVQLVIPQLLNFWLHLMFFYFFMFIIILNLWRVTSI 629
Query: 418 SQLNLTFREIIYNYRKQPQCKPQV 441
LNL F ++ N + P KPQV
Sbjct: 628 EFLNLAF--LVVNSTQHPIIKPQV 563
>BQ144385 similar to GP|21740500|emb OSJNBa0006B20.15 {Oryza sativa}, partial
(14%)
Length = 1329
Score = 28.9 bits (63), Expect = 6.2
Identities = 18/48 (37%), Positives = 23/48 (47%), Gaps = 5/48 (10%)
Frame = -2
Query: 324 PPPLQSAFASPTKPNPGGGGETCAET-----GISVFFSKIVKLGAVRI 366
PPP P P PGGG +T +T G + FS + K G VR+
Sbjct: 158 PPP-------PRLPKPGGGKKTVTQTPPARGGGAPLFSFLFKRGGVRV 36
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.342 0.148 0.505
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,837,498
Number of Sequences: 36976
Number of extensions: 274714
Number of successful extensions: 2100
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 1559
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 522
Number of HSP's gapped (non-prelim): 1657
length of query: 624
length of database: 9,014,727
effective HSP length: 102
effective length of query: 522
effective length of database: 5,243,175
effective search space: 2736937350
effective search space used: 2736937350
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146862.5


