

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146862.25 + phase: 0
(206 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
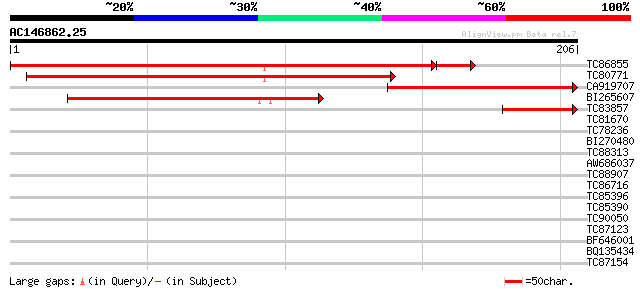
Score E
Sequences producing significant alignments: (bits) Value
TC86855 similar to GP|20466312|gb|AAM20473.1 unknown protein {Ar... 281 5e-78
TC80771 similar to GP|16612243|gb|AAL27494.1 At2g35190/T4C15.14 ... 129 8e-31
CA919707 similar to GP|19569611|gb| NPSN12 {Arabidopsis thaliana... 120 4e-28
BI265607 similar to GP|16612243|gb| At2g35190/T4C15.14 {Arabidop... 92 1e-19
TC83857 homologue to GP|16612243|gb|AAL27494.1 At2g35190/T4C15.1... 44 4e-05
TC81670 weakly similar to PIR|E86293|E86293 hypothetical protein... 35 0.026
TC78236 weakly similar to GP|15451010|gb|AAK96776.1 Unknown prot... 31 0.38
BI270480 weakly similar to GP|9757727|db contains similarity to ... 30 0.85
TC88313 weakly similar to PIR|E84776|E84776 hypothetical protein... 30 0.85
AW686037 similar to PIR|E64203|E642 ATP-dependent nuclease addA ... 29 1.4
TC88907 similar to GP|14194159|gb|AAK56274.1 At1g71270/F3I17_8 {... 29 1.4
TC86716 similar to PIR|T14279|T14279 myosin-like protein my5 - c... 28 1.9
TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protei... 28 1.9
TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 ... 28 2.5
TC90050 similar to GP|22002145|gb|AAM88629.1 putative glyoxylate... 28 2.5
TC87123 weakly similar to GP|21554031|gb|AAM63112.1 unknown {Ara... 27 4.2
BF646001 homologue to GP|20303561|gb| putative kinase {Oryza sat... 27 7.2
BQ135434 homologue to GP|7239240|gb|A ICP27 {human herpesvirus 1... 27 7.2
TC87154 similar to GP|2289893|gb|AAB65076.1| unknown {Saccharomy... 26 9.4
>TC86855 similar to GP|20466312|gb|AAM20473.1 unknown protein {Arabidopsis
thaliana}, partial (73%)
Length = 846
Score = 281 bits (718), Expect(2) = 5e-78
Identities = 151/185 (81%), Positives = 153/185 (82%), Gaps = 30/185 (16%)
Frame = +2
Query: 1 MASNLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIK 60
MASNLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIK
Sbjct: 248 MASNLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIK 427
Query: 61 DFDREIKDEGAGNPEEVNKQLNDEKQSMTEK----------------------------- 91
DFDREIKDEGAGNPEEVNKQLNDEKQSM ++
Sbjct: 428 DFDREIKDEGAGNPEEVNKQLNDEKQSMIKELNSYVALRKTYMNTIGNKKLELFDMGAGA 607
Query: 92 -NSTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQT 150
STAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQT
Sbjct: 608 SESTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQT 787
Query: 151 EQMGR 155
EQMGR
Sbjct: 788 EQMGR 802
Score = 27.3 bits (59), Expect(2) = 5e-78
Identities = 13/14 (92%), Positives = 13/14 (92%)
Frame = +3
Query: 156 IVNELDSIQFSIKK 169
I NELDSIQFSIKK
Sbjct: 804 IGNELDSIQFSIKK 845
>TC80771 similar to GP|16612243|gb|AAL27494.1 At2g35190/T4C15.14
{Arabidopsis thaliana}, partial (58%)
Length = 701
Score = 129 bits (324), Expect = 8e-31
Identities = 72/161 (44%), Positives = 99/161 (60%), Gaps = 27/161 (16%)
Frame = +1
Query: 7 MTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIKDFDREI 66
++ L +I G + D FRAL+NGFQKL+KIKD++R+S QLEELT KMR+CK LIK+FD+E+
Sbjct: 217 ISEDLAEIDGRVADNFRALSNGFQKLEKIKDASRKSRQLEELTEKMRECKGLIKEFDKEV 396
Query: 67 KDEGAGNPEEVNKQLNDEKQSMTEK---------------------------NSTAEGNV 99
K A E K LN++KQSM ++ E NV
Sbjct: 397 KALEASFDRETTKFLNEKKQSMIKELNSYVALKKQYASNIENKRIELFEGPNEGDTEENV 576
Query: 100 QLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGT 140
LAS MSN++L++ G + MD+TDQAIER K++V T+ +GT
Sbjct: 577 LLASAMSNEQLMDHGNRMMDDTDQAIER*KRIVQDTVNIGT 699
>CA919707 similar to GP|19569611|gb| NPSN12 {Arabidopsis thaliana}, partial
(36%)
Length = 659
Score = 120 bits (301), Expect = 4e-28
Identities = 64/69 (92%), Positives = 65/69 (93%)
Frame = -1
Query: 138 VGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCG 197
V TQTAST KGQTEQMGRIVNEL IQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCG
Sbjct: 659 VCTQTAST*KGQTEQMGRIVNELIQIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCG 480
Query: 198 VIAIIVVKV 206
VIAIIVVK+
Sbjct: 479 VIAIIVVKI 453
>BI265607 similar to GP|16612243|gb| At2g35190/T4C15.14 {Arabidopsis
thaliana}, partial (26%)
Length = 416
Score = 92.4 bits (228), Expect = 1e-19
Identities = 57/121 (47%), Positives = 73/121 (60%), Gaps = 28/121 (23%)
Frame = +3
Query: 22 FRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIKDFDREIKDEGAGNPEEVNKQL 81
FRAL+NGFQKL+KIKD++R+S QLEELT KMR+CK LIK+FD+E+K A E K L
Sbjct: 45 FRALSNGFQKLEKIKDASRKSRQLEELTEKMRECKGLIKEFDKEVKALEASFDRETTKFL 224
Query: 82 NDEKQSMT-EKNS---------------------------TAEGNVQLASEMSNQELVNA 113
N++KQSM E NS T + NV LAS MSN++L++
Sbjct: 225 NEKKQSMIKELNSYVALKKQYRFKY*E*AN*TL*GXPMKVTQKXNVLLASAMSNEQLMDH 404
Query: 114 G 114
G
Sbjct: 405 G 407
>TC83857 homologue to GP|16612243|gb|AAL27494.1 At2g35190/T4C15.14
{Arabidopsis thaliana}, partial (17%)
Length = 610
Score = 43.9 bits (102), Expect = 4e-05
Identities = 21/27 (77%), Positives = 24/27 (88%)
Frame = +3
Query: 180 QVATDKCIMLFLFLIVCGVIAIIVVKV 206
+VATDKCIM LFLIV GVIAII+VK+
Sbjct: 48 KVATDKCIMALLFLIVIGVIAIIIVKL 128
>TC81670 weakly similar to PIR|E86293|E86293 hypothetical protein AAF18488.1
[imported] - Arabidopsis thaliana, partial (41%)
Length = 1797
Score = 34.7 bits (78), Expect = 0.026
Identities = 27/104 (25%), Positives = 52/104 (49%), Gaps = 1/104 (0%)
Frame = +2
Query: 27 NGFQKLDKIKDSNRQ-STQLEELTGKMRDCKRLIKDFDREIKDEGAGNPEEVNKQLNDEK 85
N +L+K D+ ++ ++EEL GK++ K L D IK + EE+N +L D+
Sbjct: 1250 NKILQLEKQLDAKQKLEMEIEELRGKLQVMKHLGDQDDTAIKKK----MEEMNSELEDKI 1417
Query: 86 QSMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSK 129
+S+ + S + + SN EL A + ++ ++ + +K
Sbjct: 1418 ESLEDMES-MNSTLIVKERQSNDELQEARKELIEGLNEMLTGAK 1546
>TC78236 weakly similar to GP|15451010|gb|AAK96776.1 Unknown protein
{Arabidopsis thaliana}, partial (61%)
Length = 1303
Score = 30.8 bits (68), Expect = 0.38
Identities = 21/77 (27%), Positives = 36/77 (46%)
Frame = +1
Query: 31 KLDKIKDSNRQSTQLEELTGKMRDCKRLIKDFDREIKDEGAGNPEEVNKQLNDEKQSMTE 90
K+ + D+ R TQL ++++ +++ +E+K E K+L DEK +
Sbjct: 451 KVSLLNDAVRVVTQLRNEAERLKERNDELREKVKELKAE--------KKELRDEKNKLKL 606
Query: 91 KNSTAEGNVQLASEMSN 107
E V+LAS SN
Sbjct: 607 DKEKLEQQVKLASVQSN 657
>BI270480 weakly similar to GP|9757727|db contains similarity to guanylate
binding protein~gene_id:MCL19.12 {Arabidopsis thaliana},
partial (12%)
Length = 657
Score = 29.6 bits (65), Expect = 0.85
Identities = 23/116 (19%), Positives = 57/116 (48%), Gaps = 7/116 (6%)
Frame = +3
Query: 44 QLEELTGKMRDCKRLIKDFDRE--IKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQL 101
++++LT +M+ + ++RE + + + E+ + + + E+ TAE
Sbjct: 108 EIKDLTDRMKSENAKAQSYEREAIVYQQEKNHLEQKYQSEFKRFEEVQERCKTAEKEAAR 287
Query: 102 ASEMSNQELVNAGMKTMDETDQ---AIERSKQV--VHQTIEVGTQTASTLKGQTEQ 152
A+E++++ AGM ++++ A+ER Q+ + IE + L+G+ ++
Sbjct: 288 ATEVADRARAEAGMAQKEKSEMQRLAMERLAQIERAERRIETLGREKDNLEGELQR 455
>TC88313 weakly similar to PIR|E84776|E84776 hypothetical protein At2g36070
[imported] - Arabidopsis thaliana, partial (31%)
Length = 1201
Score = 29.6 bits (65), Expect = 0.85
Identities = 20/97 (20%), Positives = 41/97 (41%)
Frame = +2
Query: 58 LIKDFDREIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKT 117
+ +F ++KDE NP E K + + K+ E EG ++ ++ + ++L
Sbjct: 233 VFNEFSNKVKDETVKNP-EFQKSVKELKEKAEELKGVKEG-LKEKTKQTTEQLYKQFDGV 406
Query: 118 MDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMG 154
E + A ++ V + I T+ G+ + G
Sbjct: 407 WKEAEAAAKKVSHNVKEKISAATEEVKAGIGKQDSSG 517
>AW686037 similar to PIR|E64203|E642 ATP-dependent nuclease addA homolog -
Mycoplasma genitalium, partial (2%)
Length = 682
Score = 28.9 bits (63), Expect = 1.4
Identities = 19/97 (19%), Positives = 42/97 (42%)
Frame = +1
Query: 58 LIKDFDREIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKT 117
+ +F +++KDE NP E + + + K+ E EG ++ ++ + ++L
Sbjct: 148 VFNEFSKKVKDETLRNP-EFQQSVKELKEKAEELKGVKEG-LKEKTKQTTEQLYRQFDGM 321
Query: 118 MDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMG 154
E + A ++ V + I T+ G+ + G
Sbjct: 322 WKEAEDAAKKVSHNVREKISAATEEVKVGIGKQDSSG 432
>TC88907 similar to GP|14194159|gb|AAK56274.1 At1g71270/F3I17_8 {Arabidopsis
thaliana}, partial (51%)
Length = 1343
Score = 28.9 bits (63), Expect = 1.4
Identities = 21/73 (28%), Positives = 37/73 (49%), Gaps = 2/73 (2%)
Frame = +1
Query: 104 EMSNQELVNAGMKTMDETDQAIERSK--QVVHQTIEVGTQTASTLKGQTEQMGRIVNELD 161
+++ +E +N ++D Q +E K +VV + G + KG + ++ ELD
Sbjct: 256 DLTIEEDLNGDDISLDGLQQELEECKNDEVVANILSKGPKLRDYTKGVENDLRKV--ELD 429
Query: 162 SIQFSIKKASQLV 174
SIQ IK++ LV
Sbjct: 430 SIQDYIKESDNLV 468
>TC86716 similar to PIR|T14279|T14279 myosin-like protein my5 - common
sunflower, partial (38%)
Length = 2675
Score = 28.5 bits (62), Expect = 1.9
Identities = 34/146 (23%), Positives = 59/146 (40%), Gaps = 10/146 (6%)
Frame = +2
Query: 44 QLEELTGKMRDCKRLIKDFDREIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLAS 103
++EELT +++ KRL D + E E A +L D +M + A V +
Sbjct: 14 RVEELTWRLQIEKRLRTDLEEEKAQEVA--------KLRDALHAMQIQVEEANAKV-IKE 166
Query: 104 EMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTL----------KGQTEQM 153
++Q+ + + ET IE ++++ EV S L + Q E
Sbjct: 167 REASQKAIQDAPPVIKETPVIIEDTEKINSLLAEVNCLKESLLLEREAKEEAKRAQAETE 346
Query: 154 GRIVNELDSIQFSIKKASQLVKEIGR 179
R ++ S +KA QL + + R
Sbjct: 347 ARSKELFKKVEDSDRKADQLQELVQR 424
>TC85396 homologue to GP|6911551|emb|CAB72129.1 heat shock protein 70
{Cucumis sativus}, complete
Length = 2320
Score = 28.5 bits (62), Expect = 1.9
Identities = 20/100 (20%), Positives = 44/100 (44%), Gaps = 16/100 (16%)
Frame = +2
Query: 59 IKDFDREIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLAS--------------- 103
+++F R+ K + +GNP + + +++ +STA+ +++ S
Sbjct: 848 VQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGVDFYTTITRAR 1027
Query: 104 -EMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQT 142
E N +L M+ +++ + + K VH + VG T
Sbjct: 1028FEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGST 1147
>TC85390 homologue to SP|P27322|HS72_LYCES Heat shock cognate 70 kDa protein
2. [Tomato] {Lycopersicon esculentum}, complete
Length = 2278
Score = 28.1 bits (61), Expect = 2.5
Identities = 21/106 (19%), Positives = 45/106 (41%), Gaps = 16/106 (15%)
Frame = +1
Query: 53 RDCKRLIKDFDREIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLAS--------- 103
R +++F R+ K + +GNP + + +++ +STA+ +++ S
Sbjct: 859 RMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDFYT 1038
Query: 104 -------EMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQT 142
E N +L M+ +++ + + K VH + VG T
Sbjct: 1039TITRARFEELNMDLFRKCMEPVEKCLRDAKMDKNSVHDVVLVGGST 1176
>TC90050 similar to GP|22002145|gb|AAM88629.1 putative glyoxylate reductase
{Oryza sativa (japonica cultivar-group)}, partial (16%)
Length = 801
Score = 28.1 bits (61), Expect = 2.5
Identities = 38/170 (22%), Positives = 73/170 (42%), Gaps = 15/170 (8%)
Frame = +1
Query: 34 KIKDSNRQSTQLEELTGKMRDCKRLIKDFDREIKDEGAGNPEEVNKQLNDEKQSMTE--- 90
++ D N QS Q + + ++ RE D+G NPE K++ + K+S ++
Sbjct: 169 EVDDENEQSDQQYK--------ENALQIIVREQLDDGYSNPESSQKKVGEVKESSSQHQV 324
Query: 91 ------KNSTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTAS 144
++ +EG +S+ + + + K+ ++DQ S++ T GT A
Sbjct: 325 SSLTLSTSTRSEGRRSRSSKKAKKRHIR--QKSQQKSDQKEGTSQR--DDTAMSGTDQA- 489
Query: 145 TLKGQTEQMGRIVNELDSIQFSIKKASQLVKEIGR------QVATDKCIM 188
L +E ++S+Q I A+Q +K R ++ D CI+
Sbjct: 490 -LSSSSEDSRSRKTPVESLQEPI--ATQALKSSTRLSGKCTELLKDGCII 630
>TC87123 weakly similar to GP|21554031|gb|AAM63112.1 unknown {Arabidopsis
thaliana}, partial (59%)
Length = 1088
Score = 27.3 bits (59), Expect = 4.2
Identities = 17/62 (27%), Positives = 28/62 (44%)
Frame = +2
Query: 98 NVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIV 157
NV L + + A M+ T + VHQ +++G + + TLKG+ RI+
Sbjct: 347 NVVLLLTAAKETTATAKMEHKKATTRKNTSFACRVHQHVKLGPKLSETLKGKLSLGARII 526
Query: 158 NE 159
E
Sbjct: 527 QE 532
>BF646001 homologue to GP|20303561|gb| putative kinase {Oryza sativa
(japonica cultivar-group)}, partial (6%)
Length = 667
Score = 26.6 bits (57), Expect = 7.2
Identities = 23/100 (23%), Positives = 47/100 (47%), Gaps = 3/100 (3%)
Frame = +2
Query: 65 EIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQA 124
E++ E N EE+ K + ++ +STAE ++ + +E+V E+
Sbjct: 179 ELETENIKNNEEIEKMKTEIEKLRKNSDSTAE--LEKEAARLRREVV--------ESKVE 328
Query: 125 IERSKQVVHQ---TIEVGTQTASTLKGQTEQMGRIVNELD 161
IE+ ++++ + IE+ + LK + +M V EL+
Sbjct: 329 IEKLRKIIDEKENKIEIVEKEGKELKQENVEMEMKVRELE 448
>BQ135434 homologue to GP|7239240|gb|A ICP27 {human herpesvirus 1}, partial
(4%)
Length = 1263
Score = 26.6 bits (57), Expect = 7.2
Identities = 21/83 (25%), Positives = 34/83 (40%), Gaps = 7/83 (8%)
Frame = -3
Query: 52 MRDCKRLIKDFDREIKDEGAGNPEEVNKQLNDEKQSMTE-------KNSTAEGNVQLASE 104
MRDC+R +++ D + + G+ + K E+ M + K A+ N
Sbjct: 616 MRDCERRVREMDMDHR----GDMRRMRKDQRTERSKMGKWTPGKIGKTGEAKPN---GKR 458
Query: 105 MSNQELVNAGMKTMDETDQAIER 127
N++L N GM M T R
Sbjct: 457 KGNEKLGNGGMGRMGLTSNRRRR 389
>TC87154 similar to GP|2289893|gb|AAB65076.1| unknown {Saccharomyces
cerevisiae}, partial (1%)
Length = 1787
Score = 26.2 bits (56), Expect = 9.4
Identities = 14/52 (26%), Positives = 23/52 (43%)
Frame = -3
Query: 61 DFDREIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVN 112
DFD + D +PE Q + + S E +Q + E+ N +L+N
Sbjct: 270 DFDDDFGDFKDASPETRFAQESTQNTSFNHPTEFNENGLQTSLEVLNSDLIN 115
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.130 0.339
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,730,700
Number of Sequences: 36976
Number of extensions: 31115
Number of successful extensions: 200
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 197
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 198
length of query: 206
length of database: 9,014,727
effective HSP length: 92
effective length of query: 114
effective length of database: 5,612,935
effective search space: 639874590
effective search space used: 639874590
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146862.25


