

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146856.7 + phase: 0
(241 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
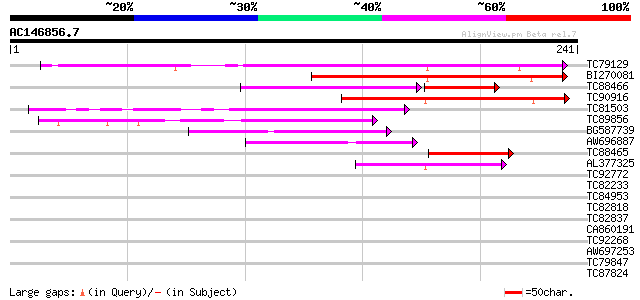
Score E
Sequences producing significant alignments: (bits) Value
TC79129 similar to PIR|A96725|A96725 hypothetical protein F20P5.... 122 9e-29
BI270081 similar to GP|6448480|emb cyclin D1 {Antirrhinum majus}... 97 7e-21
TC88466 similar to GP|6434199|emb|CAB60837.1 CycD3;2 {Lycopersic... 74 1e-20
TC90916 similar to PIR|T09961|T09961 cyclin D-like protein - red... 81 3e-16
TC81503 similar to GP|6448480|emb|CAB61221.1 cyclin D1 {Antirrhi... 81 3e-16
TC89856 homologue to GP|4583990|emb|CAB40540.1 cyclin D3 {Medica... 69 2e-12
BG587739 GP|18147003|db cyclin D2 {Sus scrofa}, partial (3%) 53 9e-08
AW696887 similar to PIR|T07669|T07 cyclin a1-type mitosis-speci... 48 4e-06
TC88465 similar to GP|6434201|emb|CAB60838.1 CycD3;3 {Lycopersic... 42 3e-04
AL377325 homologue to GP|3608181|dbj cyclin B {Pisum sativum}, p... 40 6e-04
TC92772 similar to GP|19347837|gb|AAL86330.1 putative prolyl end... 39 0.002
TC82233 homologue to PIR|T09598|T09598 cyclin 4 D-type - alfalf... 38 0.004
TC84953 weakly similar to PIR|T39562|T39562 60S ribosomal protei... 33 0.076
TC82818 similar to GP|20259203|gb|AAM14317.1 unknown protein {Ar... 30 1.1
TC82837 similar to GP|20197453|gb|AAC73041.2 expressed protein {... 28 2.4
CA860191 homologue to GP|19697342|gb hypothetical protein {Dicty... 28 3.2
TC92268 similar to GP|4803836|dbj|BAA77516.1 a dynamin-like prot... 28 4.1
AW697253 similar to GP|12597289|gb| FasL isoform {Homo sapiens},... 27 5.4
TC79847 similar to GP|21689689|gb|AAM67466.1 unknown protein {Ar... 27 5.4
TC87824 homologue to GP|16612249|gb|AAL27496.1 AT4g35230/F23E12_... 27 5.4
>TC79129 similar to PIR|A96725|A96725 hypothetical protein F20P5.7
[imported] - Arabidopsis thaliana, partial (64%)
Length = 1328
Score = 122 bits (307), Expect = 9e-29
Identities = 82/230 (35%), Positives = 124/230 (53%), Gaps = 6/230 (2%)
Frame = +2
Query: 14 LMCHEQDESTLIFEEDEDESTFFINSSLDNNNNNPWFLLDDEEEYIQYLFKQETGL--GF 71
L+C E +S+ + D E + ++SS + + ++EEE I + E GF
Sbjct: 143 LLCGE--DSSEVLTGDLPECSSDLDSSSSSQLPSSSLFAEEEEESIAVFIEHEFKFVPGF 316
Query: 72 GSITHFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNTQAKFGFTVQTAYLSINYF 131
++ F S+SL + R AI WI +GF TAYLS+NY
Sbjct: 317 DYVSRF--------------QSRSLE--SSTREEAIAWILKVHEYYGFQPLTAYLSVNYM 448
Query: 132 DRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPIE---YRFENKVIKNME 188
DRFL R + ES W +QLLSVACLS+AAKMEE VP L ++ IE Y F+ + I ME
Sbjct: 449 DRFLDSRPLPESNGWPLQLLSVACLSLAAKMEEPLVPSLLDFQIEGAKYIFQPRTILRME 628
Query: 189 LLILSTLEWKMGLPTPFAYLHYFFTKF-CNGSRSETIITKATQHIVTMVK 237
LL+L+ L+W++ TP ++L +F K G+ + II++AT+ I++ ++
Sbjct: 629 LLVLTILDWRLRSITPLSFLSFFACKLDSTGTFTHFIISRATEIILSNIQ 778
>BI270081 similar to GP|6448480|emb cyclin D1 {Antirrhinum majus}, partial
(50%)
Length = 663
Score = 96.7 bits (239), Expect = 7e-21
Identities = 51/113 (45%), Positives = 76/113 (67%), Gaps = 4/113 (3%)
Frame = +2
Query: 129 NYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPIE---YRFENKVIK 185
NY DRFL+ R + ++ W +QLLSVACLS+AAKMEE VP L + +E Y FE I+
Sbjct: 2 NYMDRFLNSRRLPQTNGWPLQLLSVACLSLAAKMEETLVPSLLDLQVEGVKYMFEPITIR 181
Query: 186 NMELLILSTLEWKMGLPTPFAYLHYFFTKFCNGSR-SETIITKATQHIVTMVK 237
MELL+LS L+W++ TPF++L +F K + S + +I++ATQ I++ ++
Sbjct: 182 RMELLVLSVLDWRLRSVTPFSFLSFFACKLDSTSTFTGFLISRATQIILSKIQ 340
>TC88466 similar to GP|6434199|emb|CAB60837.1 CycD3;2 {Lycopersicon
esculentum}, partial (39%)
Length = 692
Score = 74.3 bits (181), Expect(2) = 1e-20
Identities = 35/77 (45%), Positives = 46/77 (59%)
Frame = +2
Query: 99 LRNARLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSI 158
L AR +I+WI A + F+ T+ L++NY DRFL KPW QL +VACLS+
Sbjct: 185 LETARRESIEWILKVNAHYSFSALTSVLAVNYLDRFLFSFRFQNEKPWMTQLAAVACLSL 364
Query: 159 AAKMEEQSVPPLSEYPI 175
AAKMEE VP L + +
Sbjct: 365 AAKMEETHVPLLLDLQV 415
Score = 42.4 bits (98), Expect(2) = 1e-20
Identities = 19/32 (59%), Positives = 23/32 (71%)
Frame = +1
Query: 177 YRFENKVIKNMELLILSTLEWKMGLPTPFAYL 208
Y FE K IK ME+LILSTL WKM TP +++
Sbjct: 502 YLFEAKTIKKMEILILSTLGWKMNPATPLSFI 597
>TC90916 similar to PIR|T09961|T09961 cyclin D-like protein - red goosefoot,
partial (31%)
Length = 886
Score = 81.3 bits (199), Expect = 3e-16
Identities = 43/102 (42%), Positives = 69/102 (67%), Gaps = 5/102 (4%)
Frame = +3
Query: 142 ESKPWAIQLLSVACLSIAAKMEEQSVPPLSEYPI---EYRFENKVIKNMELLILSTLEWK 198
+ + W +QLL+VACLS+AAK+EE +VP + I ++ FE K I+ MELL+LSTL+W+
Sbjct: 18 KGRAWTMQLLAVACLSLAAKVEETAVPQPLDLQIGESKFVFEAKTIQRMELLVLSTLKWR 197
Query: 199 MGLPTPFAYLHYFFTKFCNGSRS--ETIITKATQHIVTMVKG 238
M TPF+++ F +K + +S + I+++TQ I + +KG
Sbjct: 198 MQAITPFSFIECFLSKIKDDDKSSLSSSISRSTQLISSTIKG 323
>TC81503 similar to GP|6448480|emb|CAB61221.1 cyclin D1 {Antirrhinum majus},
partial (36%)
Length = 1000
Score = 81.3 bits (199), Expect = 3e-16
Identities = 58/162 (35%), Positives = 82/162 (49%)
Frame = +3
Query: 9 FSLSSLMCHEQDESTLIFEEDEDESTFFINSSLDNNNNNPWFLLDDEEEYIQYLFKQETG 68
F+ S L+C E+ S L D T S + + P E+E+I L + +
Sbjct: 180 FTDSDLLCGEETSSIL----SSDSPT----ESFSDGESYP----PPEDEFIAGLIEDQGK 323
Query: 69 LGFGSITHFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNTQAKFGFTVQTAYLSI 128
G D+ V++ KS + +AR +I WI Q +GF TAYL++
Sbjct: 324 FVIGF--------DYFVKM------KSSSFDSDARDESIRWILKVQGYYGFQPVTAYLAV 461
Query: 129 NYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVPPL 170
NY DRFL+ R + ++ W +QLLSVACLS+AAKMEE VP L
Sbjct: 462 NYMDRFLNSRRLPQTNGWPLQLLSVACLSLAAKMEETLVPSL 587
>TC89856 homologue to GP|4583990|emb|CAB40540.1 cyclin D3 {Medicago sativa},
partial (41%)
Length = 689
Score = 68.6 bits (166), Expect = 2e-12
Identities = 48/157 (30%), Positives = 73/157 (45%), Gaps = 13/157 (8%)
Frame = +1
Query: 13 SLMCHEQ--------DESTLIFEEDEDESTFFINSS-LDNNNNNPWFLLD----DEEEYI 59
+L CHE+ DE ++ EE E N+ LD+ + P LL+ +++E +
Sbjct: 256 ALYCHEEKWEDDDNDDEGEVVDEEGEQSYVTTTNNDILDSTSLLPLLLLEQNLFNKDEEL 435
Query: 60 QYLFKQETGLGFGSITHFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNTQAKFGF 119
LF +E I YDD + D L R A++W+ A +GF
Sbjct: 436 NTLFSKE------KIQQETYYDDLKNVMNFDS-------LTQPRREAVEWMLKVNAHYGF 576
Query: 120 TVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACL 156
+ TA L++NY DRFL + KPW IQL++V C+
Sbjct: 577 SALTATLAVNYLDRFLLSFHFQKEKPWMIQLVAVTCI 687
>BG587739 GP|18147003|db cyclin D2 {Sus scrofa}, partial (3%)
Length = 519
Score = 53.1 bits (126), Expect = 9e-08
Identities = 33/86 (38%), Positives = 45/86 (51%)
Frame = +3
Query: 77 FLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFLS 136
FL DH + + KS + + R I I +Q F YL+INY DRFL+
Sbjct: 204 FLIESDHIPPLNYFQNLKSNEFDASVRTDFISLI--SQLSCNFDPFVTYLAINYLDRFLA 377
Query: 137 KRSIDESKPWAIQLLSVACLSIAAKM 162
+ I + KPWA +LL+V C S+A KM
Sbjct: 378 NQGILQPKPWANKLLAVTCFSLAVKM 455
>AW696887 similar to PIR|T07669|T07 cyclin a1-type mitosis-specific -
soybean, partial (33%)
Length = 648
Score = 47.8 bits (112), Expect = 4e-06
Identities = 25/73 (34%), Positives = 41/73 (55%)
Frame = +3
Query: 101 NARLHAIDWIFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAA 160
N R +DW+ ++ +T +LS++Y DRFLS + SK +QLL V+ + IA+
Sbjct: 435 NMRGTLVDWLVEVADEYKLLPETLHLSVSYIDRFLSIEPVSRSK---LQLLGVSSMLIAS 605
Query: 161 KMEEQSVPPLSEY 173
K EE + P ++
Sbjct: 606 KYEEITPPKAVDF 644
>TC88465 similar to GP|6434201|emb|CAB60838.1 CycD3;3 {Lycopersicon
esculentum}, partial (30%)
Length = 637
Score = 41.6 bits (96), Expect = 3e-04
Identities = 18/36 (50%), Positives = 24/36 (66%)
Frame = +3
Query: 179 FENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTK 214
FE K IK ME+LILSTL WKM TP +++ + +
Sbjct: 9 FEAKTIKKMEILILSTLGWKMNPATPLSFIDFIIRR 116
>AL377325 homologue to GP|3608181|dbj cyclin B {Pisum sativum}, partial (45%)
Length = 329
Score = 40.4 bits (93), Expect = 6e-04
Identities = 21/66 (31%), Positives = 39/66 (58%), Gaps = 2/66 (3%)
Frame = +2
Query: 148 IQLLSVACLSIAAKMEEQSVPPLSEYPI--EYRFENKVIKNMELLILSTLEWKMGLPTPF 205
+QL+ ++ + +A+K EE P ++++ + + ++ I ME IL LEW + +PTPF
Sbjct: 14 LQLVGISAMLMASKYEEIWPPEVNDFVCLSDRAYSHEQILIMEKTILGKLEWTLTVPTPF 193
Query: 206 AYLHYF 211
+L F
Sbjct: 194 VFLVRF 211
>TC92772 similar to GP|19347837|gb|AAL86330.1 putative prolyl endopeptidase
{Arabidopsis thaliana}, partial (14%)
Length = 531
Score = 38.5 bits (88), Expect = 0.002
Identities = 19/63 (30%), Positives = 37/63 (58%), Gaps = 2/63 (3%)
Frame = +3
Query: 148 IQLLSVACLSIAAKMEEQSVPPLSEYPI--EYRFENKVIKNMELLILSTLEWKMGLPTPF 205
+QL+ ++ + IA+K EE P ++++ + + + + ME IL LEW + +PTP+
Sbjct: 333 LQLVGISSMLIASKYEEIWAPEVNDFVCISDNAYVREQVLVMEKTILRNLEWYLTVPTPY 512
Query: 206 AYL 208
+L
Sbjct: 513 VFL 521
>TC82233 homologue to PIR|T09598|T09598 cyclin 4 D-type - alfalfa, partial
(51%)
Length = 1011
Score = 37.7 bits (86), Expect = 0.004
Identities = 18/36 (50%), Positives = 24/36 (66%)
Frame = +1
Query: 179 FENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTK 214
FE K I+ MELLILSTL+WKM T ++L + +
Sbjct: 4 FEAKTIQRMELLILSTLKWKMHPVTTHSFLDHIIRR 111
>TC84953 weakly similar to PIR|T39562|T39562 60S ribosomal protein L32 -
fission yeast (Schizosaccharomyces pombe), partial (86%)
Length = 442
Score = 33.5 bits (75), Expect = 0.076
Identities = 22/79 (27%), Positives = 40/79 (49%), Gaps = 3/79 (3%)
Frame = -3
Query: 55 EEEYIQYLFKQETGLGFGSITHFLCYDDHDVEVEDDDSSKSLF---WLRNARLHAIDWIF 111
++E +Q + +E+GLG GS+T D D V + SS S+ WL A ++ +
Sbjct: 224 DQESLQSVGSEESGLGVGSVT-----DGDDWLVSSESSSHSVIDTSWLSPAWSQSVIKVA 60
Query: 112 NTQAKFGFTVQTAYLSINY 130
+ G ++ A+L +N+
Sbjct: 59 RESGELGSSLVEAFLLLNW 3
>TC82818 similar to GP|20259203|gb|AAM14317.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 1176
Score = 29.6 bits (65), Expect = 1.1
Identities = 26/112 (23%), Positives = 48/112 (42%), Gaps = 2/112 (1%)
Frame = -2
Query: 110 IFNTQAKFGFTVQTAYLSINYFDRFLSKRSIDESKPWAIQLLSVACLSIAAKMEE--QSV 167
+ + A F ++T Y +N + + + D K + + ++ L+ A K + + V
Sbjct: 1016 VMDNHALISFPIKTVYFQLN-----IPQITKDIRKLYIVHVMP---LTKACKQKT*LKLV 861
Query: 168 PPLSEYPIEYRFENKVIKNMELLILSTLEWKMGLPTPFAYLHYFFTKFCNGS 219
PP+S +RF +K I++ + ILS F L+ FC S
Sbjct: 860 PPIS-----FRFSSKAIRSFSIFILSFSSLNFSNSRSFFLLNLSHLNFCFSS 720
>TC82837 similar to GP|20197453|gb|AAC73041.2 expressed protein {Arabidopsis
thaliana}, partial (52%)
Length = 913
Score = 28.5 bits (62), Expect = 2.4
Identities = 15/34 (44%), Positives = 20/34 (58%), Gaps = 3/34 (8%)
Frame = +3
Query: 17 HEQDESTLI---FEEDEDESTFFINSSLDNNNNN 47
+E D ST++ EDEDE +S DNN+NN
Sbjct: 114 NEDDTSTMLSPSHNEDEDEIEDAADSDADNNDNN 215
>CA860191 homologue to GP|19697342|gb hypothetical protein {Dictyostelium
discoideum}, partial (1%)
Length = 504
Score = 28.1 bits (61), Expect = 3.2
Identities = 12/21 (57%), Positives = 16/21 (76%)
Frame = +2
Query: 27 EEDEDESTFFINSSLDNNNNN 47
EE+EDE T NS+ +NNN+N
Sbjct: 272 EEEEDEFTTTFNSNNNNNNSN 334
Score = 27.3 bits (59), Expect = 5.4
Identities = 14/35 (40%), Positives = 22/35 (62%)
Frame = +2
Query: 13 SLMCHEQDESTLIFEEDEDESTFFINSSLDNNNNN 47
+L E++E EEDE +TF N++ +N+NNN
Sbjct: 236 TLQNDEEEEEEDEEEEDEFTTTFNSNNNNNNSNNN 340
>TC92268 similar to GP|4803836|dbj|BAA77516.1 a dynamin-like protein ADL3
{Arabidopsis thaliana}, partial (35%)
Length = 1034
Score = 27.7 bits (60), Expect = 4.1
Identities = 38/176 (21%), Positives = 68/176 (38%), Gaps = 8/176 (4%)
Frame = +1
Query: 1 MGSYVGTPFSLSSLMCHEQDESTL------IFEEDEDESTFFINSSLDNNNNNPWFLLDD 54
MG + G +QD+ T + D + + ++ D WF+L++
Sbjct: 481 MGQHSGGNLKSMKEKSGQQDKDTQEVSSLKVAGPDGEITAGYLFKKSDKGWTKRWFVLNE 660
Query: 55 EEEYIQYLFKQETGLGFGSITHFLCYDDHDVEVEDDDSSKSLFWLRNARLHAIDWIFNTQ 114
+ + Y KQE G+IT C D ++ ED+ +KS ++ + + +D +
Sbjct: 661 KSGKLGYTKKQEERHFRGTITLEECNID-EISDEDEAPAKS---SKDKKSNGLDSGKASN 828
Query: 115 AKFGFTVQTAYLSINYFDR--FLSKRSIDESKPWAIQLLSVACLSIAAKMEEQSVP 168
F T + Y +I L S+ + W +L VA + E S P
Sbjct: 829 LIFKITSKVPYKTIMKAQSTVLLKAESMVDKAEWINKLRKVAQAKGGQAIGEPSFP 996
>AW697253 similar to GP|12597289|gb| FasL isoform {Homo sapiens}, partial
(16%)
Length = 761
Score = 27.3 bits (59), Expect = 5.4
Identities = 11/22 (50%), Positives = 14/22 (63%)
Frame = +2
Query: 27 EEDEDESTFFINSSLDNNNNNP 48
EE+E+E I +NNNNNP
Sbjct: 143 EEEEEEEVRIIRRRRNNNNNNP 208
>TC79847 similar to GP|21689689|gb|AAM67466.1 unknown protein {Arabidopsis
thaliana}, partial (44%)
Length = 1504
Score = 27.3 bits (59), Expect = 5.4
Identities = 12/41 (29%), Positives = 21/41 (50%)
Frame = +1
Query: 59 IQYLFKQETGLGFGSITHFLCYDDHDVEVEDDDSSKSLFWL 99
I + Q + + FG+ C+ ++D ++ DDD L WL
Sbjct: 1381 IMFFLSQISSVSFGTANLLYCHMNND-DMNDDDDINILIWL 1500
>TC87824 homologue to GP|16612249|gb|AAL27496.1 AT4g35230/F23E12_210
{Arabidopsis thaliana}, partial (36%)
Length = 895
Score = 27.3 bits (59), Expect = 5.4
Identities = 11/22 (50%), Positives = 14/22 (63%)
Frame = +1
Query: 27 EEDEDESTFFINSSLDNNNNNP 48
EE+E+E I +NNNNNP
Sbjct: 151 EEEEEEEVRIIRRRRNNNNNNP 216
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,622,179
Number of Sequences: 36976
Number of extensions: 131740
Number of successful extensions: 764
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 745
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 754
length of query: 241
length of database: 9,014,727
effective HSP length: 93
effective length of query: 148
effective length of database: 5,575,959
effective search space: 825241932
effective search space used: 825241932
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146856.7


