

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146856.6 - phase: 0 /pseudo
(335 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
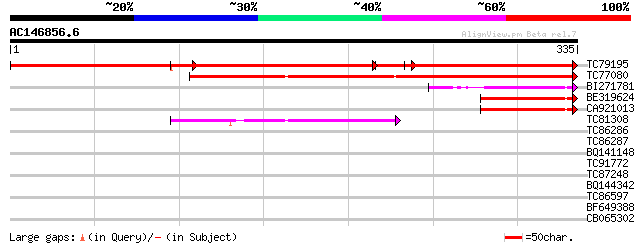
Score E
Sequences producing significant alignments: (bits) Value
TC79195 similar to GP|15529178|gb|AAK97683.1 AT5g67480/K9I9_4 {A... 250 e-133
TC77080 similar to GP|10177297|dbj|BAB10558. contains similarity... 219 1e-57
BI271781 homologue to GP|9954734|gb Unknown Protein {Arabidopsis... 67 1e-11
BE319624 60 2e-09
CA921013 similar to GP|12597461|gb p300/CBP acetyltransferase-re... 60 2e-09
TC81308 similar to PIR|T01838|T01838 hypothetical protein T15F16... 55 4e-08
TC86286 similar to GP|6862923|gb|AAF30312.1| unknown protein {Ar... 33 0.12
TC86287 similar to GP|23617108|dbj|BAC20790. contains ESTs AU056... 32 0.26
BQ141148 homologue to EGAD|147836|157 hypothetical protein 87B1.... 30 1.0
TC91772 weakly similar to GP|17380998|gb|AAL36311.1 unknown prot... 30 1.7
TC87248 similar to GP|7594580|emb|CAB88075.1 hypothetical protei... 30 1.7
BQ144342 28 6.5
TC86597 weakly similar to GP|9837280|gb|AAG00510.1| leaf senesce... 28 6.5
BF649388 28 6.5
CB065302 similar to PIR|B83246|B83 probable transcription regula... 27 8.5
>TC79195 similar to GP|15529178|gb|AAK97683.1 AT5g67480/K9I9_4 {Arabidopsis
thaliana}, partial (78%)
Length = 1519
Score = 250 bits (639), Expect(3) = e-133
Identities = 122/122 (100%), Positives = 122/122 (100%)
Frame = +2
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL 155
MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL
Sbjct: 413 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHL 592
Query: 156 KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQS 215
KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQS
Sbjct: 593 KRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQS 772
Query: 216 HP 217
HP
Sbjct: 773 HP 778
Score = 224 bits (572), Expect(3) = e-133
Identities = 100/102 (98%), Positives = 102/102 (99%)
Frame = +1
Query: 234 KKERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDFKANQPCRYTSCKGLELLV 293
++ERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDFKANQPCRYTSCKGLELLV
Sbjct: 829 RRERIRKMNEREVYLQLYDAMEALVHICRDGCRTIGPHDKDFKANQPCRYTSCKGLELLV 1008
Query: 294 RHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
RHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC
Sbjct: 1009RHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 1134
Score = 188 bits (478), Expect = 2e-48
Identities = 97/116 (83%), Positives = 105/116 (89%), Gaps = 5/116 (4%)
Frame = +1
Query: 1 MQSSEA*ADG*HGWKITPEC*EIHSDTTSFASISSNRLSLRVCGDE*FITERAKQLCLFC 60
++SSEA*ADG*HGWKITPEC*EIHSDTTSFASISSNRLSLRVCGDE*FITERAKQLCLFC
Sbjct: 103 IKSSEA*ADG*HGWKITPEC*EIHSDTTSFASISSNRLSLRVCGDE*FITERAKQLCLFC 282
Query: 61 NKRSVGASFQ*RV*SRCLH*Y**RWHYLCSF*HY-----CMLKQANRSNRWRSISI 111
NKRSVGASFQ*RV*SRCLH*Y**RWHYLCSF*HY C+ + A S ++S++I
Sbjct: 283 NKRSVGASFQ*RV*SRCLH*Y**RWHYLCSF*HYCGGFSCIERHAEASKSFQSVAI 450
Score = 40.4 bits (93), Expect(3) = e-133
Identities = 20/24 (83%), Positives = 21/24 (87%)
Frame = +3
Query: 217 PVLEKEILESMIDEENSKKERIRK 240
PVLEKEILESMIDEENSKK +K
Sbjct: 777 PVLEKEILESMIDEENSKKGEDQK 848
>TC77080 similar to GP|10177297|dbj|BAB10558. contains similarity to unknown
protein~gene_id:MDC12.13~pir||T06706 {Arabidopsis
thaliana}, partial (67%)
Length = 1539
Score = 219 bits (558), Expect = 1e-57
Identities = 115/233 (49%), Positives = 154/233 (65%), Gaps = 4/233 (1%)
Frame = +2
Query: 107 RSISIFGVPHDAVRVFIRYLYSSCYEKEEMKEFVLHLLVLSHVYAVPHLKRECEQKLELG 166
R I I GVP AV F+ +LYSS ++EM ++ +HLL LSHVY VP LK+ C + L +
Sbjct: 302 RIIQIHGVPSTAVTAFVGFLYSSSCTEDEMDKYGIHLLALSHVYNVPQLKQRCIKGL-VQ 478
Query: 167 LLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMKQSHPVLEKEILES 226
LT +N+VDV QLA LCDAP L + C + + +F V ++EGWK + + PVLE +IL
Sbjct: 479 RLTSENVVDVLQLARLCDAPDLRVRCMKLLTNHFNAVQKTEGWKFLTKHDPVLELDILRF 658
Query: 227 MIDEENSKKERIRKMNERE-VYLQLYDAMEALVHICRDGCRTIGP-HDKDFKANQPC-RY 283
M DE +++E RK E + +Y +L +AME L HIC +GC +GP H + K +PC +Y
Sbjct: 659 M-DESETRRENSRKHREEQGLYAELSEAMECLEHICTEGCTDVGPYHVEVNKEKKPCSKY 835
Query: 284 TSCKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADP-DFCRVPLC 335
++C+GL+LL+RHFA CK RV GGC CKRMWQL LHS +C D C VPLC
Sbjct: 836 STCQGLQLLIRHFATCKKRVKGGCWRCKRMWQLFRLHSYVCQQTHDSCNVPLC 994
>BI271781 homologue to GP|9954734|gb Unknown Protein {Arabidopsis thaliana},
partial (7%)
Length = 655
Score = 66.6 bits (161), Expect = 1e-11
Identities = 36/88 (40%), Positives = 50/88 (55%)
Frame = +2
Query: 248 LQLYDAMEALVHICRDGCRTIGPHDKDFKANQPCRYTSCKGLELLVRHFAGCKLRVPGGC 307
LQL ++ LVH + CR+ PH C+Y +C+ ++ L RH CK R GGC
Sbjct: 29 LQLRKMLDLLVHASQ--CRS--PH---------CQYPNCRKVKGLFRHGMHCKTRASGGC 169
Query: 308 GHCKRMWQLLELHSRLCADPDFCRVPLC 335
CK+MW LL+LH+R C + + C VP C
Sbjct: 170 VLCKKMWYLLQLHARACKESE-CHVPRC 250
>BE319624
Length = 531
Score = 59.7 bits (143), Expect = 2e-09
Identities = 24/57 (42%), Positives = 37/57 (64%)
Frame = +2
Query: 279 QPCRYTSCKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
+PC + + ++ L H + CK+R+ GGC HCK++W +L HSR C D + CR+P C
Sbjct: 128 EPCSHPN*SEIKKLFYHASKCKIRLNGGCQHCKKIWFILTSHSRNCKDSE-CRIPRC 295
>CA921013 similar to GP|12597461|gb p300/CBP acetyltransferase-related
protein 2 {Arabidopsis thaliana}, partial (1%)
Length = 780
Score = 59.7 bits (143), Expect = 2e-09
Identities = 24/57 (42%), Positives = 37/57 (64%)
Frame = -2
Query: 279 QPCRYTSCKGLELLVRHFAGCKLRVPGGCGHCKRMWQLLELHSRLCADPDFCRVPLC 335
+PC + + ++ L H + CK+R+ GGC HCK++W +L HSR C D + CR+P C
Sbjct: 536 EPCSHPN*SEIKKLFYHASKCKIRLNGGCQHCKKIWFILTSHSRNCKDSE-CRIPRC 369
>TC81308 similar to PIR|T01838|T01838 hypothetical protein T15F16.14 -
Arabidopsis thaliana, partial (57%)
Length = 1141
Score = 55.1 bits (131), Expect = 4e-08
Identities = 39/138 (28%), Positives = 71/138 (51%), Gaps = 2/138 (1%)
Frame = +2
Query: 96 MLKQANRSNRWRSISIFGVPHDAVRVFIRYLYSS--CYEKEEMKEFVLHLLVLSHVYAVP 153
ML+ +R +I I V +DA+R F+ YLY++ C + + +LLVL Y V
Sbjct: 536 MLENDMEESRSGTIKIADVSYDALRAFVNYLYTAEACLDNQ----MACNLLVLGEKYQVK 703
Query: 154 HLKRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILKNFRTVSESEGWKAMK 213
HLK CE+ L + L + + F A +A +L I++N ++++E + +
Sbjct: 704 HLKAYCEKYL-ISKLNWEKAIVNFAFAHQHNANQLQDAALAVIMENMDNLTKNEDYTELV 880
Query: 214 QSHPVLEKEILESMIDEE 231
+++P L EI E+ + ++
Sbjct: 881 ETNPRLVVEIYEAYLAKQ 934
>TC86286 similar to GP|6862923|gb|AAF30312.1| unknown protein {Arabidopsis
thaliana}, partial (91%)
Length = 1729
Score = 33.5 bits (75), Expect = 0.12
Identities = 24/86 (27%), Positives = 44/86 (50%), Gaps = 2/86 (2%)
Frame = +3
Query: 142 HLLVLSHVYAVPHLKRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKIL--KN 199
HLL + YA+ L+ CE L + ++ + LA +L +C + I +N
Sbjct: 1155 HLLAAADRYALERLRLICEASL-CEDVAINTVATTLALAEQHHCFQLKAVCLKFIATSEN 1331
Query: 200 FRTVSESEGWKAMKQSHPVLEKEILE 225
R V +++G++ +K+S P + E+LE
Sbjct: 1332 LRAVMQTDGFEYLKESCPSVLTELLE 1409
>TC86287 similar to GP|23617108|dbj|BAC20790. contains ESTs AU056959(S21019)
AU056960(S21019)~similar to Arabidopsis thaliana
chromosome 5, partial (82%)
Length = 1302
Score = 32.3 bits (72), Expect = 0.26
Identities = 23/85 (27%), Positives = 43/85 (50%), Gaps = 2/85 (2%)
Frame = +3
Query: 142 HLLVLSHVYAVPHLKRECEQKLELGLLTMDNLVDVFQLALLCDAPRLSLICHRKILK--N 199
HLL + YA+ L+ CE L + ++ + LA +L +C + I + N
Sbjct: 1050 HLLAAADRYALDRLRLMCEASL-CEDVAINTVATTLALAEQHHCFQLKAVCLKFIARPEN 1226
Query: 200 FRTVSESEGWKAMKQSHPVLEKEIL 224
R V +++G++ +K+S P + E+L
Sbjct: 1227 LRAVMQTDGFEYLKESCPSVLTELL 1301
>BQ141148 homologue to EGAD|147836|157 hypothetical protein 87B1.h
{Drosophila melanogaster}, partial (4%)
Length = 485
Score = 30.4 bits (67), Expect = 1.0
Identities = 13/27 (48%), Positives = 18/27 (66%), Gaps = 1/27 (3%)
Frame = -3
Query: 285 SCKGLELLVRHFAGCKLRVPGG-CGHC 310
+C GL+L++R G + VPGG C HC
Sbjct: 108 ACAGLDLVLRQPWGARAEVPGGACHHC 28
>TC91772 weakly similar to GP|17380998|gb|AAL36311.1 unknown protein
{Arabidopsis thaliana}, partial (17%)
Length = 1237
Score = 29.6 bits (65), Expect = 1.7
Identities = 19/53 (35%), Positives = 28/53 (51%)
Frame = +2
Query: 194 RKILKNFRTVSESEGWKAMKQSHPVLEKEILESMIDEENSKKERIRKMNEREV 246
R + + + S S+ K K+S+ V EKE E D ++SKKE +NE V
Sbjct: 161 RSVRRKGKKASSSKSTKPSKKSNVVSEKEA-EKTADSKSSKKEVPISLNEDSV 316
>TC87248 similar to GP|7594580|emb|CAB88075.1 hypothetical protein
{Arabidopsis thaliana}, partial (82%)
Length = 1271
Score = 29.6 bits (65), Expect = 1.7
Identities = 24/92 (26%), Positives = 44/92 (47%), Gaps = 2/92 (2%)
Frame = +1
Query: 161 QKLELGLLTMDNLVDVFQLALLCDAP--RLSLICHRKILKNFRTVSESEGWKAMKQSHPV 218
+KL+ L+ + + + + L D RL+ ++L N+R ES WK + +
Sbjct: 298 EKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNELLPNWRPTMESLFWKILSAGKEL 477
Query: 219 LEKEILESMIDEENSKKERIRKMNEREVYLQL 250
L L +DE + E+I +N+ E +L+L
Sbjct: 478 LSLIALSLNLDE--NYFEKISALNKPEAFLRL 567
>BQ144342
Length = 799
Score = 27.7 bits (60), Expect = 6.5
Identities = 15/40 (37%), Positives = 20/40 (49%)
Frame = -3
Query: 191 ICHRKILKNFRTVSESEGWKAMKQSHPVLEKEILESMIDE 230
+CH R V E W A+ +SH L KE++ S DE
Sbjct: 254 LCHSPFTA-MRHVREG*SWVAILESHARLAKEVITSNSDE 138
>TC86597 weakly similar to GP|9837280|gb|AAG00510.1| leaf
senescence-associated receptor-like protein kinase
{Phaseolus vulgaris}, partial (46%)
Length = 2233
Score = 27.7 bits (60), Expect = 6.5
Identities = 13/30 (43%), Positives = 16/30 (53%)
Frame = -2
Query: 75 SRCLH*Y**RWHYLCSF*HYCMLKQANRSN 104
S CLH Y * WH +C+ C+ N SN
Sbjct: 264 SSCLHQYL*IWHLICNL---CLSSMKNSSN 184
>BF649388
Length = 349
Score = 27.7 bits (60), Expect = 6.5
Identities = 10/39 (25%), Positives = 24/39 (60%), Gaps = 1/39 (2%)
Frame = +1
Query: 119 VRVFIRY-LYSSCYEKEEMKEFVLHLLVLSHVYAVPHLK 156
++++I++ ++SC+ + K+++L L H Y P +K
Sbjct: 211 IKLYIKFNFHTSCFSFPKKKKYLLFXXFLKHTYICPFVK 327
>CB065302 similar to PIR|B83246|B83 probable transcription regulator PA3215
[imported] - Pseudomonas aeruginosa (strain PAO1),
partial (36%)
Length = 461
Score = 27.3 bits (59), Expect = 8.5
Identities = 17/54 (31%), Positives = 25/54 (45%), Gaps = 4/54 (7%)
Frame = -1
Query: 264 GCRTIGP--HDKDFKANQPCRYTSCKGLELLVRHFAGCKLRVPGG--CGHCKRM 313
G R +G H + A+ C GL V+ +AG LR PG HC+++
Sbjct: 353 GGRAVGQRRHRPEHGASGTSSLVPCGGLRADVQSYAGRGLRAPGALPAHHCRKL 192
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.342 0.149 0.512
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,943,092
Number of Sequences: 36976
Number of extensions: 215582
Number of successful extensions: 1480
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 1466
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1478
length of query: 335
length of database: 9,014,727
effective HSP length: 97
effective length of query: 238
effective length of database: 5,428,055
effective search space: 1291877090
effective search space used: 1291877090
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146856.6


