

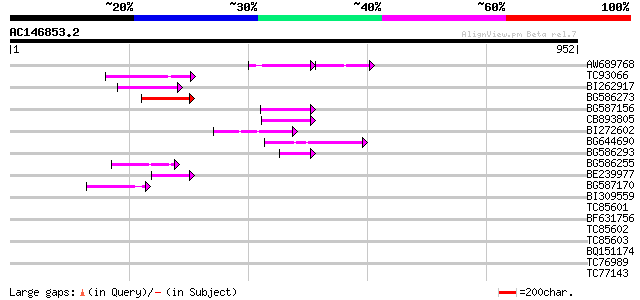
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146853.2 - phase: 0 /pseudo
(952 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 89 4e-24
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 89 6e-18
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 84 2e-16
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 80 5e-15
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 67 4e-11
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 67 4e-11
BI272602 GP|11602812|db avenaIII {Gallus gallus}, partial (3%) 59 7e-09
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 59 1e-08
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 50 4e-06
BG586255 similar to GP|7682800|gb Hypothetical protein T15F17.l ... 50 4e-06
BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryz... 49 7e-06
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 45 1e-04
BI309559 homologue to GP|437310|gb|A nodulin {Medicago truncatul... 41 0.002
TC85601 nodulin 41 0.002
BF631756 41 0.002
TC85602 similar to GP|437310|gb|AAA62850.1|| nodulin {Medicago t... 40 0.006
TC85603 similar to GP|437310|gb|AAA62850.1|| nodulin {Medicago t... 40 0.006
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 39 0.007
TC76989 homologue to SP|O24310|EFTU_PEA Elongation factor Tu ch... 38 0.016
TC77143 similar to GP|559557|gb|AAA51649.1|| arabinogalactan-pro... 38 0.021
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 89.4 bits (220), Expect(2) = 4e-24
Identities = 49/113 (43%), Positives = 61/113 (53%), Gaps = 1/113 (0%)
Frame = +1
Query: 402 ITRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVSA-LKDPNWKMAMNDEYNSLIKNKTWDL 460
ITR++ G KP P P++ + L DP W AM EY +LI NKTWDL
Sbjct: 1 ITRTKTGKLKPKV--------FLTEPCPQDCENLPLSDPRWLQAMKTEYKALIDNKTWDL 156
Query: 461 VPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
VP PP I W++R KE DG+ + KARLV G Q +G D ETFSP+
Sbjct: 157 VPLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVAKGFSQTLGCDYTETFSPV 315
Score = 41.2 bits (95), Expect(2) = 4e-24
Identities = 39/99 (39%), Positives = 47/99 (47%)
Frame = +2
Query: 514 *NQLLFAQFSELLHPDLGKFISWMSRMLFFMVN*KKQCTCTNLWDLKILKTPIMYAVFAN 573
*N L FS LL GKF +S M F MV K++C C NL DLK+L + A N
Sbjct: 317 *NLSLSGSFSPLLSLINGKFNKLISTMPF*MVFFKRKCICLNLRDLKLL-INLWCAS*TN 493
Query: 574 HSMGLNKLLELGISDLSTTCHLLVFLKANVITLFSYTRK 612
HSM +K G+ LV LK VI + T K
Sbjct: 494 HSMA*SKHHVHGMRX*PVHKFSLVSLKVVVIHHYLSTIK 610
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 89.4 bits (220), Expect = 6e-18
Identities = 53/155 (34%), Positives = 81/155 (52%), Gaps = 4/155 (2%)
Frame = +1
Query: 161 NSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPS--SFWHHAL 218
+S+F + C +GI + P QNG +ER IR+ R +L +A L + W A
Sbjct: 325 SSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAA 504
Query: 219 EMATYLMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKC 278
A +L+N PH ++F+ P + Y++LR+FGC + L KL PR+ +C
Sbjct: 505 STACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYALVNDG---KLAPRAGEC 675
Query: 279 VFLGYPSNHRGYK--CLGLLSNKIIICRHVLFNEN 311
+FL Y S +GY+ C S K+I+ R V FNE+
Sbjct: 676 IFLSYASESKGYRLWCSDPKSQKLILSRDVTFNED 780
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 84.3 bits (207), Expect = 2e-16
Identities = 41/110 (37%), Positives = 64/110 (57%)
Frame = +1
Query: 181 YTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATYLMNILPHKTINFESPLK 240
+T QNG +ER R+ R +L A + SFW A++ A Y++N P I+ ++P++
Sbjct: 82 HTPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPME 261
Query: 241 VLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLGYPSNHRGY 290
+ K Y+ L VFGC + ++ S + KL P+S KC+FLGY N +GY
Sbjct: 262 MWKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGY 411
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 79.7 bits (195), Expect = 5e-15
Identities = 35/90 (38%), Positives = 56/90 (61%)
Frame = -2
Query: 221 ATYLMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVF 280
A YL+N +P + + ++P +VL + PS ++RVFGCLC+ L P KL+ RS K +F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 281 LGYPSNHRGYKCLGLLSNKIIICRHVLFNE 310
+GY + +GYKC + ++++ R V F E
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIE 435
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 66.6 bits (161), Expect = 4e-11
Identities = 34/92 (36%), Positives = 51/92 (54%)
Frame = -1
Query: 422 HVTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEK 481
++ ++ +PR+ A++D WK ++ E ++IKN TW P + S WIF K K
Sbjct: 579 NLNENHIPRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWIFTIKYK 400
Query: 482 SDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
+DG+ ER K RLV G G D ETF+P+
Sbjct: 399 ADGSIERKKTRLVARGFTLTYGEDYIETFAPV 304
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 66.6 bits (161), Expect = 4e-11
Identities = 32/91 (35%), Positives = 48/91 (52%)
Frame = +3
Query: 423 VTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKS 482
+T + P A+K W+ +MN+E + +N TW+L I WIF+ K
Sbjct: 24 LTMTSDPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNE 203
Query: 483 DGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
+G E++KARLV G QQ G+D E F+P+
Sbjct: 204 NGEIEKYKARLVAKGYSQQYGVDYTEVFAPV 296
>BI272602 GP|11602812|db avenaIII {Gallus gallus}, partial (3%)
Length = 488
Score = 59.3 bits (142), Expect = 7e-09
Identities = 47/146 (32%), Positives = 68/146 (46%), Gaps = 6/146 (4%)
Frame = +1
Query: 343 DQTQTGPPIPQPAH---QPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQANT 399
+ TQT P ++ Q I+SP + N + ++PT P PP +P T
Sbjct: 4 ESTQTSSPSSPNSNEVAQNIQISSPASPTSNELANITTSPTPPPPP--PPTARPHNNHTT 177
Query: 400 KP---ITRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKNK 456
ITRSQ+ I K +K LH P N AL+D +W+ M DE+++L N
Sbjct: 178 NVGGIITRSQNNIVKTIKKL-NLHVRPLCPIEPSNITQALRDHDWRSIMQDEFDALQNNN 354
Query: 457 TWDLVPRPPDVNVIRSMWIFRHKEKS 482
TWDLV R N++ F ++ KS
Sbjct: 355 TWDLVSRSFAQNLVGCKLGFSYQTKS 432
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 58.5 bits (140), Expect = 1e-08
Identities = 50/173 (28%), Positives = 79/173 (44%)
Frame = -3
Query: 429 PRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFER 488
P+N AL+D +W +M +E + ++K W LVPRP VI + W+FR+ K + E
Sbjct: 585 PKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVFRN--KLE*LLEI 412
Query: 489 HKARLVGDGAGQQVGIDCGETFSPM*NQLLFAQFSELLHPDLGKFISWMSRMLFFMVN*K 548
+ + D ++ F + L LLH WM R+ M K
Sbjct: 411 NPSWWCKDTI-KKKE*TMMRLFHLLPEWKLLEF**LLLHSWGSSCTKWM*RVHLLMEISK 235
Query: 549 KQCTCTNLWDLKILKTPIMYAVFANHSMGLNKLLELGISDLSTTCHLLVFLKA 601
++C +NL DLK+ + IM + H M +KL E G+ + C +V +A
Sbjct: 234 RRCLSSNLLDLKMQRYQIMCSD*IRHYMV*SKLQEHGMKGCQSFC*RMVSKEA 76
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 50.1 bits (118), Expect = 4e-06
Identities = 25/60 (41%), Positives = 35/60 (57%)
Frame = +2
Query: 454 KNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
+ +T LV +P V I WI++ K DGT ++KARLV G +Q GID E F+P+
Sbjct: 47 QKQTLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPV 226
>BG586255 similar to GP|7682800|gb Hypothetical protein T15F17.l {Arabidopsis
thaliana}, partial (12%)
Length = 436
Score = 50.1 bits (118), Expect = 4e-06
Identities = 34/115 (29%), Positives = 58/115 (49%), Gaps = 1/115 (0%)
Frame = -1
Query: 172 GIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATYLMNILPHK 231
G V+ S + +QNG +E I+ I R LL+ + P S W HA+ A L+ I P
Sbjct: 415 G*VWNTSVVHVHTQNGLAESFIKRIQLITRPLLMRSKQPVSAWGHAVLHAAELIRIRPSS 236
Query: 232 TINFESPLKVLYHKDPSYNHLRVFGCLCF-PLFPSSKIYKLQPRSTKCVFLGYPS 285
+ SP ++L P +H++ FG + P+ P + K+ P+ +++G+ S
Sbjct: 235 EHKY-SPSQLLSGHVPDVSHIKTFGYPVYVPIAPPHRT-KMGPQRRMGIYVGFDS 77
>BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 514
Score = 49.3 bits (116), Expect = 7e-06
Identities = 28/74 (37%), Positives = 37/74 (49%), Gaps = 2/74 (2%)
Frame = -2
Query: 239 LKVLYHKDPSYNHL--RVFGCLCFPLFPSSKIYKLQPRSTKCVFLGYPSNHRGYKCLGLL 296
L Y P+ N+L R+FGC F S K R+ KCVF+ Y S +GY+C
Sbjct: 504 LPSFYPYVPTSNNLTPRIFGCTSFVHIHSDGRSKFDHRALKCVFIRYSSTQKGYRCYHPP 325
Query: 297 SNKIIICRHVLFNE 310
S K + R V F+E
Sbjct: 324 SRKYFVSRDVTFHE 283
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 45.1 bits (105), Expect = 1e-04
Identities = 29/109 (26%), Positives = 53/109 (48%), Gaps = 1/109 (0%)
Frame = -3
Query: 129 KFIQYLKN*EHTSTHNFKEKLKQFNVTMAVNVNSN-FQKMCEANGIVFRLSCPYTSSQNG 187
+ I KN + T+++ K+K S F+ + +GI+ + SCPYT QNG
Sbjct: 305 RVIDAFKNFQAYVTNHYHAKIKILRSDNGGEYTSYAFKSHLDHHGILHQTSCPYTPQQNG 126
Query: 188 KSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATYLMNILPHKTINFE 236
++RK + + R+L+ A++ + A YL+N +P K + +
Sbjct: 125 VAKRKNKHLMEVARSLMFQANVST---------ACYLINWIPTKVLRIK 6
>BI309559 homologue to GP|437310|gb|A nodulin {Medicago truncatula}, partial
(38%)
Length = 644
Score = 40.8 bits (94), Expect = 0.002
Identities = 27/89 (30%), Positives = 45/89 (50%), Gaps = 4/89 (4%)
Frame = +3
Query: 349 PPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQ---ANTKPITRS 405
PP+ P HI P + P + PP +P PI++ PPI++P ++ A P+
Sbjct: 42 PPVENPQFYKPHIEKPPVHKPLVEKPPTHHP--PIEK--PPIYKPPVEKPPAYKPPV--E 203
Query: 406 QHGIFKPNQKYYGLH-THVTKSPLPRNPV 433
H ++KP+ + +H V K P+ + PV
Sbjct: 204 HHPVYKPSVEKPPVHKPPVEKPPVHKPPV 290
Score = 32.0 bits (71), Expect = 1.2
Identities = 22/88 (25%), Positives = 41/88 (46%), Gaps = 3/88 (3%)
Frame = +3
Query: 349 PPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQP---TLQANTKPITRS 405
PP+ +P H+ P + P + PP P P+++ PP+ +P L + P+ +
Sbjct: 312 PPVEKPPVYKPHVEKPPLHKPPVEKPPVHKP--PVEK--PPVHKPPVEKLPVHKPPVEKP 479
Query: 406 QHGIFKPNQKYYGLHTHVTKSPLPRNPV 433
++KP + +H K P+ + PV
Sbjct: 480 P--VYKPPVEKPPVH----KPPVEKPPV 545
>TC85601 nodulin
Length = 1933
Score = 40.8 bits (94), Expect = 0.002
Identities = 27/89 (30%), Positives = 45/89 (50%), Gaps = 4/89 (4%)
Frame = +3
Query: 349 PPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQ---ANTKPITRS 405
PP+ P HI P + P + PP +P PI++ PPI++P ++ A P+
Sbjct: 102 PPVENPQFYKPHIEKPPVHKPLVEKPPTHHP--PIEK--PPIYKPPVEKPPAYKPPV--E 263
Query: 406 QHGIFKPNQKYYGLH-THVTKSPLPRNPV 433
H ++KP+ + +H V K P+ + PV
Sbjct: 264 HHPVYKPSVEKPPVHKPPVEKPPVHKPPV 350
Score = 35.4 bits (80), Expect = 0.10
Identities = 24/89 (26%), Positives = 44/89 (48%), Gaps = 4/89 (4%)
Frame = +3
Query: 349 PPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQANTKPITR---S 405
PP+ +P H+ P N P + PP P P+++ PP+ +P ++ P+ +
Sbjct: 612 PPVEKPPVYKPHVEKPPVNKPPVEKPPVHKP--PVEK--PPVHKPPVE--KLPVYKPHVE 773
Query: 406 QHGIFKPNQKYYGLH-THVTKSPLPRNPV 433
+ ++KP + LH V K+P+ + PV
Sbjct: 774 KPPVYKPLVEKPPLHKPPVEKTPMHKPPV 860
Score = 34.3 bits (77), Expect = 0.23
Identities = 24/88 (27%), Positives = 39/88 (44%), Gaps = 3/88 (3%)
Frame = +3
Query: 349 PPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQP---TLQANTKPITRS 405
PP+ +P H+ P + P + PP P P+++ PP+ +P L P+ +
Sbjct: 927 PPVEKPPVYKPHVEKPPLHKPPVEKPPVHKP--PVEK--PPVHKPPVEKLPVYKPPVEKP 1094
Query: 406 QHGIFKPNQKYYGLHTHVTKSPLPRNPV 433
++KP HV K PL + PV
Sbjct: 1095P--VYKP---------HVEKPPLHKPPV 1145
Score = 30.4 bits (67), Expect = 3.4
Identities = 22/87 (25%), Positives = 37/87 (42%), Gaps = 2/87 (2%)
Frame = +3
Query: 349 PPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQANT--KPITRSQ 406
PP+ +P H+ P + P + PP P P+++ PP+ +P ++ KP
Sbjct: 1077 PPVEKPPVYKPHVEKPPLHKPPVEKPPVHKP--PVEK--PPVHKPPVEKPPVHKPPVEKL 1244
Query: 407 HGIFKPNQKYYGLHTHVTKSPLPRNPV 433
P +K V K P+ + PV
Sbjct: 1245 PVYKPPVEKPPVYKPPVEKPPVHKPPV 1325
Score = 29.3 bits (64), Expect = 7.5
Identities = 23/88 (26%), Positives = 42/88 (47%), Gaps = 3/88 (3%)
Frame = +3
Query: 349 PPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQANT--KPITRSQ 406
PP+ +P + P + P + PP P P+++ PP+ +P ++ KP + +
Sbjct: 312 PPVEKPPVHKPPVEKPPVHKPPVEKPPVHKP--PVEK--PPVHKPPVEKLPVYKP-SVEK 476
Query: 407 HGIFKPNQKYYGLHTH-VTKSPLPRNPV 433
++KP + LH V K P+ + PV
Sbjct: 477 PPVYKPPVEKPPLHKPLVEKPPVHKPPV 560
>BF631756
Length = 622
Score = 40.8 bits (94), Expect = 0.002
Identities = 15/26 (57%), Positives = 20/26 (76%)
Frame = +2
Query: 436 LKDPNWKMAMNDEYNSLIKNKTWDLV 461
L+DP W +AMN EY++L+ N TW LV
Sbjct: 545 LRDPKWHLAMNTEYDTLLANNTWSLV 622
>TC85602 similar to GP|437310|gb|AAA62850.1|| nodulin {Medicago truncatula},
partial (48%)
Length = 833
Score = 39.7 bits (91), Expect = 0.006
Identities = 26/89 (29%), Positives = 42/89 (46%), Gaps = 4/89 (4%)
Frame = +1
Query: 349 PPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQ---ANTKPITRS 405
PP+ P HI P + P + PP +P Q PPI++P ++ A P+
Sbjct: 58 PPVENPQFYKPHIEKPPVHKPLVEKPPTHHP----QIEKPPIYKPPVEKPPAYKPPV--E 219
Query: 406 QHGIFKPNQKYYGLH-THVTKSPLPRNPV 433
H ++KP+ + +H V K P+ + PV
Sbjct: 220 HHPVYKPSVEKPPVHKPPVDKPPVHKPPV 306
Score = 30.8 bits (68), Expect = 2.6
Identities = 26/107 (24%), Positives = 46/107 (42%), Gaps = 3/107 (2%)
Frame = +1
Query: 349 PPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQP---TLQANTKPITRS 405
PP+ +P + P + P + PP P P+++ PP+ +P L + P+ +
Sbjct: 388 PPVEKPPVYKPQVEKPPLHKPPVEKPPVHKP--PVEK--PPVHKPPVEKLPVHKPPVEKP 555
Query: 406 QHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEYNSL 452
++KP + +H + P P S + N A N E +SL
Sbjct: 556 P--VYKPPVEKPPVHKPPVEKPPVHKPSSG--EAN*IQATN*EVSSL 684
>TC85603 similar to GP|437310|gb|AAA62850.1|| nodulin {Medicago truncatula},
partial (70%)
Length = 1183
Score = 39.7 bits (91), Expect = 0.006
Identities = 26/89 (29%), Positives = 42/89 (46%), Gaps = 4/89 (4%)
Frame = +3
Query: 349 PPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQ---ANTKPITRS 405
PP+ P HI P + P + PP +P Q PPI++P ++ A P+
Sbjct: 99 PPVENPQFYKPHIEKPPVHKPLVEKPPTHHP----QIEKPPIYKPPVEKPPAYKPPV--E 260
Query: 406 QHGIFKPNQKYYGLH-THVTKSPLPRNPV 433
H ++KP+ + +H V K P+ + PV
Sbjct: 261 HHPVYKPSVEKPPVHKPPVDKPPVHKPPV 347
Score = 35.0 bits (79), Expect = 0.14
Identities = 24/86 (27%), Positives = 42/86 (47%), Gaps = 1/86 (1%)
Frame = +3
Query: 349 PPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTLQANTKPITRSQHG 408
PP+ +P H+ P + P + PP P P+++ PP+ +P ++ P+ +
Sbjct: 894 PPVEKPPVYKPHVEKPPLHKPPVEKPPVHKP--PVEK--PPVHKPPVE--KLPVEKLP-- 1049
Query: 409 IFKPN-QKYYGLHTHVTKSPLPRNPV 433
++KP +K HV K PL + PV
Sbjct: 1050VYKPPVEKPPVYKPHVEKPPLHKPPV 1127
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 39.3 bits (90), Expect = 0.007
Identities = 28/95 (29%), Positives = 37/95 (38%), Gaps = 3/95 (3%)
Frame = +2
Query: 349 PPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQ---PTLQANTKPITRS 405
PP PQP P +P P +PN P ++ P ++ PP P + P
Sbjct: 11 PPNPQPPSDPPKKRTPKPETPNRKKPQKNKKEPPKKRKTPPEASHPPPRPRPPRPPQPNH 190
Query: 406 QHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPN 440
QH KP + + H H T P P P PN
Sbjct: 191 QHD--KPQKTTHPHHPH-TPPPTPHPPNPPPNHPN 286
Score = 30.4 bits (67), Expect = 3.4
Identities = 19/54 (35%), Positives = 24/54 (44%), Gaps = 4/54 (7%)
Frame = +3
Query: 347 TGPPIPQPAHQPNHITSPCPNSPNITSPPQ----SNPTSPIQQNLPPIFQPTLQ 396
T PP P P PN T N T+P + + PT P + + PP PT Q
Sbjct: 144 TPPPAPAPPGPPNQTT-------NTTNPKKQHTPTTPTHPPRPHTPPTPPPTTQ 284
Score = 29.3 bits (64), Expect = 7.5
Identities = 12/29 (41%), Positives = 14/29 (47%)
Frame = +3
Query: 354 PAHQPNHITSPCPNSPNITSPPQSNPTSP 382
P H P T P P T+PP+ PT P
Sbjct: 231 PTHPPRPHTPPTPPPTTQTTPPREAPTDP 317
>TC76989 homologue to SP|O24310|EFTU_PEA Elongation factor Tu chloroplast
precursor (EF-Tu). [Garden pea] {Pisum sativum}, partial
(92%)
Length = 1907
Score = 38.1 bits (87), Expect = 0.016
Identities = 25/71 (35%), Positives = 32/71 (44%), Gaps = 5/71 (7%)
Frame = +1
Query: 340 HLMDQTQTGPPIPQPAHQPNHITSPCPNSPNITSPPQS-----NPTSPIQQNLPPIFQPT 394
H T T P+P P P PN P +TSPP S + TSP+ N PP+ P
Sbjct: 193 HPTHHTLTPSPLPLPLPFPTPPPLIVPN*P*LTSPPPSSTPPQSSTSPLPPNPPPL-APP 369
Query: 395 LQANTKPITRS 405
L + P+ S
Sbjct: 370 LSPSVPPVANS 402
>TC77143 similar to GP|559557|gb|AAA51649.1|| arabinogalactan-protein {Pyrus
communis}, partial (30%)
Length = 956
Score = 37.7 bits (86), Expect = 0.021
Identities = 22/59 (37%), Positives = 24/59 (40%), Gaps = 2/59 (3%)
Frame = +3
Query: 349 PPIPQPAHQPNHITSPCPNSPNITSPPQSNPT--SPIQQNLPPIFQPTLQANTKPITRS 405
PP P P P T P P T PP + PT +P PP PT A T T S
Sbjct: 222 PPTPAPVATPPTATPPTATPPTATPPPAAAPTPATPAPATSPPAPTPTSDAPTPDSTSS 398
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.337 0.147 0.487
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,527,640
Number of Sequences: 36976
Number of extensions: 685525
Number of successful extensions: 8747
Number of sequences better than 10.0: 182
Number of HSP's better than 10.0 without gapping: 4287
Number of HSP's successfully gapped in prelim test: 531
Number of HSP's that attempted gapping in prelim test: 2766
Number of HSP's gapped (non-prelim): 5894
length of query: 952
length of database: 9,014,727
effective HSP length: 105
effective length of query: 847
effective length of database: 5,132,247
effective search space: 4347013209
effective search space used: 4347013209
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146853.2


