

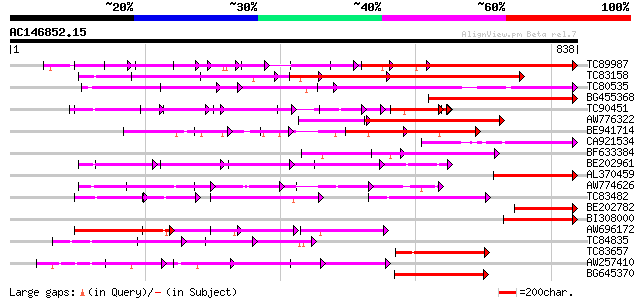
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146852.15 + phase: 0
(838 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89987 weakly similar to PIR|D86443|D86443 probable PPR-repeat ... 307 e-102
TC83158 weakly similar to PIR|H86191|H86191 hypothetical protein... 273 3e-73
TC80535 weakly similar to GP|14335136|gb|AAK59848.1 At2g15690/F9... 199 3e-51
BG455368 weakly similar to GP|9759524|dbj selenium-binding prote... 191 1e-48
TC90451 similar to PIR|H84508|H84508 hypothetical protein At2g13... 139 1e-46
AW776322 weakly similar to PIR|H84859|H84 hypothetical protein A... 162 4e-40
BE941714 weakly similar to GP|11994734|dbj selenium-binding prot... 158 9e-39
CA921534 similar to PIR|D96516|D96 F16N3.14 [imported] - Arabido... 148 7e-36
BF633384 similar to PIR|T46179|T46 hypothetical protein T8H10.30... 142 5e-34
BE202961 weakly similar to GP|6016735|gb|A hypothetical protein ... 135 6e-32
AL370459 weakly similar to GP|20146256|d hypothetical protein {O... 132 5e-31
AW774626 similar to PIR|T47909|T47 hypothetical protein T20K12.7... 130 2e-30
TC83482 weakly similar to GP|9758453|dbj|BAB08982.1 selenium-bin... 130 3e-30
BE202782 weakly similar to GP|22093801|dbj hypothetical protein~... 127 2e-29
BI308000 similar to GP|7363286|dbj| Similar to Arabidopsis thali... 124 2e-28
AW696172 weakly similar to PIR|H96620|H96 hypothetical protein F... 115 7e-26
TC84835 weakly similar to PIR|T02328|T02328 hypothetical protein... 113 3e-25
TC83657 weakly similar to GP|11994382|dbj|BAB02341. gb|AAB82628.... 110 2e-24
AW257410 weakly similar to GP|10177533|db selenium-binding prote... 109 5e-24
BG645370 weakly similar to PIR|T00405|T004 hypothetical protein ... 108 8e-24
>TC89987 weakly similar to PIR|D86443|D86443 probable PPR-repeat protein
[imported] - Arabidopsis thaliana, partial (62%)
Length = 1559
Score = 307 bits (787), Expect(2) = e-102
Identities = 143/318 (44%), Positives = 218/318 (67%)
Frame = +2
Query: 521 MRVFENMEKRDVSAWTAAIRVKAVEGNAKGAIELFDEMLKQDVKADDFVFVALLTAFSHG 580
+RVF+NM +++ ++T I A+ G K A+++F EM+++ + DD V+V + +A SH
Sbjct: 575 LRVFKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSACSHA 754
Query: 581 GYVDQGRQLFWAMEKIHGVSPQIVHYGCMVDLLGRAGLLEEAFDLMKSMPIKPNDVIWGS 640
G V++G Q F +M+ H + P + HYGCMVDLLGR G+L+EA++L+KSM IKPNDVIW S
Sbjct: 755 GLVEEGLQCFKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEAYELIKSMSIKPNDVIWRS 934
Query: 641 FLAACRKHKNVEFANYADEKITQLAPEKVGIHVLLSNIYASAGKWNDVARVRLQMKEKGF 700
L+AC+ H N+E A E + L G +++L+N+YA A KW+DVA++R ++ E+
Sbjct: 935 LLSACKVHHNLEIGKIAAENLFMLNQNNSGDYLVLANMYAKAQKWDDVAKIRTKLAERNL 1114
Query: 701 QKVAGSSSIEVHGLIREFTSGDESHTENAQIGLMLQEINCRISQVGYVPDTTNVLVDVDE 760
+ G S IE + +F S D+S + I M+ ++ ++ GY+PDT+ VL+DVD+
Sbjct: 1115VQTPGFSLIEAKRKVYKFVSQDKSIPQWNIIYEMIHQMEWQVKFEGYIPDTSQVLLDVDD 1294
Query: 761 QEKEHLLSRHSEKLAMAYGLINTGKGIPIRVVKNLRMCSDCHSFAKLVSKLYGREITVRD 820
+EK+ L HS+KLA+A+GLI+T +G P+R+ +NLRMCSDCH++ K +S +Y REITVRD
Sbjct: 1295EEKKERLKFHSQKLAIAFGLIHTSEGXPLRITRNLRMCSDCHTYTKYISMIYEREITVRD 1474
Query: 821 NNRYHFFKEGFCSCRDFW 838
R+ FK G CSC+D+W
Sbjct: 1475RXRFXHFKNGSCSCKDYW 1528
Score = 96.3 bits (238), Expect = 4e-20
Identities = 54/143 (37%), Positives = 81/143 (55%), Gaps = 1/143 (0%)
Frame = +1
Query: 141 ACSKIMAFSEGVQVHGVVVKMGLVKDLFVANSLIHFYAACGKVDLGRKVFDEMLERNVVS 200
ACS + EG+QVHG V KMGL D+ V NSLI+ Y CG++ VF+ M E++V S
Sbjct: 130 ACSLLGVVDEGIQVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVAS 309
Query: 201 WTSLINGYSVVNMAKEAVCLFFEMVEVG-VEPNPVTMVCAISACAKLKDLELGKKVCNLM 259
W+++I ++ V M E + L +M G T+V +SAC L +LGK + ++
Sbjct: 310 WSAIIGAHACVEMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLGKCIHGIL 489
Query: 260 TELGVKSNTLVVNALLDMYMKCG 282
+ N +V +L+DMY+K G
Sbjct: 490 LRNISELNVVVKTSLIDMYVKSG 558
Score = 82.4 bits (202), Expect(2) = e-102
Identities = 51/174 (29%), Positives = 88/174 (50%), Gaps = 1/174 (0%)
Frame = +1
Query: 343 ACAQLGDLSVGKSSHAYVFRNGLERLDNISNAIIDMYMKCGKREAACKVFDSMSNKTVVT 402
AC+ LG + G H +VF+ GLE + N++I+MY KCG
Sbjct: 130 ACSLLGVVDEGIQVHGHVFKMGLEGDVIVQNSLINMYGKCG------------------- 252
Query: 403 WNSLIAGLVRDGELELALRIFGEMPESNLVSWNTMIGAMVQASMFEEAIDLLREMQNQG- 461
E++ A +F M E ++ SW+ +IGA M+ E + LL +M ++G
Sbjct: 253 ------------EIKNACDVFNGMDEKSVASWSAIIGAHACVEMWNECLMLLGKMSSEGR 396
Query: 462 IKGDRVTMVGIASACGYLGALDLAKWIYTYIEKNDIHIDMQLGTALVDMFSRCG 515
+ + T+V + SAC +LG+ DL K I+ + +N +++ + T+L+DM+ + G
Sbjct: 397 CRVEESTLVNVLSACTHLGSPDLGKCIHGILLRNISELNVVVKTSLIDMYVKSG 558
Score = 77.0 bits (188), Expect(2) = 4e-23
Identities = 40/143 (27%), Positives = 85/143 (58%), Gaps = 1/143 (0%)
Frame = +1
Query: 242 ACAKLKDLELGKKVCNLMTELGVKSNTLVVNALLDMYMKCGDMYAVREIFDEFSDKNLVM 301
AC+ L ++ G +V + ++G++ + +V N+L++MY KCG++ ++F+ +K++
Sbjct: 130 ACSLLGVVDEGIQVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVAS 309
Query: 302 YNTIMSNYVQHGLAGEVLVVLDEMLQKGQ-RPDKVTMLSTIAACAQLGDLSVGKSSHAYV 360
++ I+ + + E L++L +M +G+ R ++ T+++ ++AC LG +GK H +
Sbjct: 310 WSAIIGAHACVEMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLGKCIHGIL 489
Query: 361 FRNGLERLDNISNAIIDMYMKCG 383
RN E + ++IDMY+K G
Sbjct: 490 LRNISELNVVVKTSLIDMYVKSG 558
Score = 58.5 bits (140), Expect = 1e-08
Identities = 39/156 (25%), Positives = 77/156 (49%), Gaps = 7/156 (4%)
Frame = +1
Query: 475 ACGYLGALDLAKWIYTYIEKNDIHIDMQLGTALVDMFSRCGDPLNAMRVFENMEKRDVSA 534
AC LG +D ++ ++ K + D+ + +L++M+ +CG+ NA VF M+++ V++
Sbjct: 130 ACSLLGVVDEGIQVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEKSVAS 309
Query: 535 WTAAIRVKAVEGNAKGAIELFDEMLKQD-VKADDFVFVALLTAFSHGGYVDQGRQLFWAM 593
W+A I A + L +M + + ++ V +L+A +H G D G+
Sbjct: 310 WSAIIGAHACVEMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLGK------ 471
Query: 594 EKIHGV------SPQIVHYGCMVDLLGRAGLLEEAF 623
IHG+ +V ++D+ ++G LE+ F
Sbjct: 472 -CIHGILLRNISELNVVVKTSLIDMYVKSGCLEKRF 576
Score = 54.3 bits (129), Expect(2) = 2e-09
Identities = 50/174 (28%), Positives = 81/174 (45%), Gaps = 19/174 (10%)
Frame = +2
Query: 186 GRKVFDEMLERNVVSWTSLINGYSVVNMAKEAVCLFFEMVEVGVEPNPVTMVCAISACAK 245
G +VF M E+N S+T +I+G ++ KEA+ +F EM+E G+ P+ V V SAC+
Sbjct: 572 GLRVFKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSACSH 751
Query: 246 LKDLELGKKVCNLMT-ELGVKSNTLVVNALLDMYMKCGDMYAVREIFDEFSDK-NLVMYN 303
+E G + M E ++ ++D+ + G + E+ S K N V++
Sbjct: 752 AGLVEEGLQCFKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEAYELIKSMSIKPNDVIWR 931
Query: 304 TIMSN-YVQHGL-----AGEVL-----------VVLDEMLQKGQRPDKVTMLST 340
+++S V H L A E L +VL M K Q+ D V + T
Sbjct: 932 SLLSACKVHHNLEIGKIAAENLFMLNQNNSGDYLVLANMYAKAQKWDDVAKIRT 1093
Score = 50.1 bits (118), Expect(2) = 4e-23
Identities = 43/160 (26%), Positives = 71/160 (43%), Gaps = 6/160 (3%)
Frame = +2
Query: 416 LELALRIFGEMPESNLVSWNTMIGAMVQASMFEEAIDLLREMQNQGIKGDRVTMVGIASA 475
L LR+F M E N S+ MI + +EA+ + EM +G+ D V VG+ SA
Sbjct: 563 LRKGLRVFKNMSEKNRYSYTVMISGLAIHGRGKEALKVFSEMIEEGLAPDDVVYVGVFSA 742
Query: 476 CGYLGALDLAKWIYTYIE-KNDIHIDMQLGTALVDMFSRCGDPLNAMRVFENME-KRDVS 533
C + G ++ + ++ ++ I +Q +VD+ R G A + ++M K +
Sbjct: 743 CSHAGLVEEGLQCFKSMQFEHKIEPTVQHYGCMVDLLGRFGMLKEAYELIKSMSIKPNDV 922
Query: 534 AWTAAIRVKAVEGNAK----GAIELFDEMLKQDVKADDFV 569
W + + V N + A LF ML Q+ D V
Sbjct: 923 IWRSLLSACKVHHNLEIGKIAAENLF--MLNQNNSGDYLV 1036
Score = 48.9 bits (115), Expect = 8e-06
Identities = 51/205 (24%), Positives = 87/205 (41%), Gaps = 1/205 (0%)
Frame = +2
Query: 97 YTCNTLIRGYAASGLCKEAIFIYLHMIIVMGIVPDNFTFPFLLSACSKIMAFSEGVQVHG 156
Y+ +I G A G KEA+ ++ MI G+ PD+ + + SACS EG+Q
Sbjct: 611 YSYTVMISGLAIHGRGKEALKVFSEMI-EEGLAPDDVVYVGVFSACSHAGLVEEGLQ--- 778
Query: 157 VVVKMGLVKDLFVANSLIHFYAACGKVDLGRKVFDEMLERNVVSWTSLINGYSVVNMAKE 216
C K F+ +E V + +++ M KE
Sbjct: 779 -----------------------CFK----SMQFEHKIEPTVQHYGCMVDLLGRFGMLKE 877
Query: 217 AVCLFFEMVE-VGVEPNPVTMVCAISACAKLKDLELGKKVCNLMTELGVKSNTLVVNALL 275
A +E+++ + ++PN V +SAC +LE+GK + L ++N+ L
Sbjct: 878 A----YELIKSMSIKPNDVIWRSLLSACKVHHNLEIGKIAAENLFMLN-QNNSGDYLVLA 1042
Query: 276 DMYMKCGDMYAVREIFDEFSDKNLV 300
+MY K V +I + +++NLV
Sbjct: 1043NMYAKAQKWDDVAKIRTKLAERNLV 1117
Score = 26.2 bits (56), Expect(2) = 2e-09
Identities = 30/136 (22%), Positives = 56/136 (41%), Gaps = 4/136 (2%)
Frame = +1
Query: 50 QLHCNMLKKG----VFNINKLIAACVQMGTHESLNYALNAFKEDEGTKCSLYTCNTLIRG 105
Q+H ++ K G V N LI + G ++ N E S+ + + +I
Sbjct: 166 QVHGHVFKMGLEGDVIVQNSLINMYGKCGEIKNACDVFNGMDEK-----SVASWSAIIGA 330
Query: 106 YAASGLCKEAIFIYLHMIIVMGIVPDNFTFPFLLSACSKIMAFSEGVQVHGVVVKMGLVK 165
+A + E + + M + T +LSAC+ + + G +HG++++
Sbjct: 331 HACVEMWNECLMLLGKMSSEGRCRVEESTLVNVLSACTHLGSPDLGKCIHGILLRNISEL 510
Query: 166 DLFVANSLIHFYAACG 181
++ V SLI Y G
Sbjct: 511 NVVVKTSLIDMYVKSG 558
>TC83158 weakly similar to PIR|H86191|H86191 hypothetical protein [imported]
- Arabidopsis thaliana, partial (42%)
Length = 1194
Score = 273 bits (697), Expect = 3e-73
Identities = 140/347 (40%), Positives = 217/347 (62%)
Frame = +2
Query: 414 GELELALRIFGEMPESNLVSWNTMIGAMVQASMFEEAIDLLREMQNQGIKGDRVTMVGIA 473
G+++ AL++F ++P N+VSW +IG V+ +EEA++ REMQ G+ D VT++ I
Sbjct: 2 GDVDDALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAII 181
Query: 474 SACGYLGALDLAKWIYTYIEKNDIHIDMQLGTALVDMFSRCGDPLNAMRVFENMEKRDVS 533
SAC LGAL L W++ + K + ++++ +L+DM++RCG A +VF+ M +R++
Sbjct: 182 SACANLGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLV 361
Query: 534 AWTAAIRVKAVEGNAKGAIELFDEMLKQDVKADDFVFVALLTAFSHGGYVDQGRQLFWAM 593
+W + I AV G A A+ F M K+ ++ + + + LTA SH G +D+G ++F +
Sbjct: 362 SWNSIIVGFAVNGLADKALSFFRSMKKEGLEPNGVSYTSALTACSHAGLIDEGLKIFADI 541
Query: 594 EKIHGVSPQIVHYGCMVDLLGRAGLLEEAFDLMKSMPIKPNDVIWGSFLAACRKHKNVEF 653
++ H SP+I HYGC+VDL RAG L+EA+D++K MP+ PN+V+ GS LAACR +VE
Sbjct: 542 KRDHRNSPRIEHYGCLVDLYSRAGRLKEAWDVIKKMPMMPNEVVLGSLLAACRTQGDVEL 721
Query: 654 ANYADEKITQLAPEKVGIHVLLSNIYASAGKWNDVARVRLQMKEKGFQKVAGSSSIEVHG 713
A + +L P +VL SNIYA+ GKW+ ++VR +MKE+G QK SSIE+
Sbjct: 722 AEKVMKYQVELYPGGDSNYVLFSNIYAAVGKWDGASKVRREMKERGLQKNLAFSSIEIDS 901
Query: 714 LIREFTSGDESHTENAQIGLMLQEINCRISQVGYVPDTTNVLVDVDE 760
I +F SGD+ H EN I L+ ++ + GYVPD + DVD+
Sbjct: 902 GIHKFVSGDKYHEENDYIYSALELLSFELHLYGYVPDFSGKESDVDD 1042
Score = 138 bits (347), Expect = 1e-32
Identities = 93/292 (31%), Positives = 155/292 (52%), Gaps = 9/292 (3%)
Frame = +2
Query: 181 GKVDLGRKVFDEMLERNVVSWTSLINGYSVVNMAKEAVCLFFEMVEVGVEPNPVTMVCAI 240
G VD K+FD++ +NVVSWT +I G+ +EA+ F EM GV P+ VT++ I
Sbjct: 2 GDVDDALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAII 181
Query: 241 SACAKLKDLELGKKVCNLMTELGVKSNTLVVNALLDMYMKCGDMYAVREIFDEFSDKNLV 300
SACA L L LG V L+ + + N V+N+L+DMY +CG + R++FD S +NLV
Sbjct: 182 SACANLGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLV 361
Query: 301 MYNTIMSNYVQHGLAGEVLVVLDEMLQKGQRPDKVTMLSTIAACAQLGDLSVGKSSHAYV 360
+N+I+ + +GLA + L M ++G P+ V+ S + AC+ G + G A +
Sbjct: 362 SWNSIIVGFAVNGLADKALSFFRSMKKEGLEPNGVSYTSALTACSHAGLIDEGLKIFADI 541
Query: 361 FRN--GLERLDNISNAIIDMYMKCGKREAACKVFDSM-SNKTVVTWNSLIAGLVRDGELE 417
R+ R+++ ++D+Y + G+ + A V M V SL+A G++E
Sbjct: 542 KRDHRNSPRIEHY-GCLVDLYSRAGRLKEAWDVIKKMPMMPNEVVLGSLLAACRTQGDVE 718
Query: 418 LALRIFGEMPE------SNLVSWNTMIGAMVQASMFEEAIDLLREMQNQGIK 463
LA ++ E SN V ++ + A+ ++ A + REM+ +G++
Sbjct: 719 LAEKVMKYQVELYPGGDSNYVLFSNIYAAV---GKWDGASKVRREMKERGLQ 865
Score = 108 bits (270), Expect = 8e-24
Identities = 83/300 (27%), Positives = 139/300 (45%), Gaps = 3/300 (1%)
Frame = +2
Query: 102 LIRGYAASGLCKEAIFIYLHMIIVMGIVPDNFTFPFLLSACSKIMAFSEGVQVHGVVVKM 161
+I G+ +EA+ + M + G+VPD T ++SAC+ + A G+ VH +V+K
Sbjct: 71 VIGGFVKKECYEEALECFREMQLA-GVVPDFVTVIAIISACANLGALGLGLWVHRLVMKK 247
Query: 162 GLVKDLFVANSLIHFYAACGKVDLGRKVFDEMLERNVVSWTSLINGYSVVNMAKEAVCLF 221
++ V NSLI YA CG ++L R+VFD M +RN+VSW S+I G++V +A +A+ F
Sbjct: 248 EFRDNVKVLNSLIDMYARCGCIELARQVFDGMSQRNLVSWNSIIVGFAVNGLADKALSFF 427
Query: 222 FEMVEVGVEPNPVTMVCAISACAKLKDLELGKKVCNLMTELGVKSNTLVVNALLDMYMKC 281
M + G+EPN V+ A++AC+ ++ G K+ +
Sbjct: 428 RSMKKEGLEPNGVSYTSALTACSHAGLIDEGLKIFADIKR-------------------- 547
Query: 282 GDMYAVREIFDEFSDKNLVMYNTIMSNYVQHGLAGEVLVVLDEMLQKGQRPDKVTMLSTI 341
D + + Y ++ Y + G E V+ +M P++V + S +
Sbjct: 548 ----------DHRNSPRIEHYGCLVDLYSRAGRLKEAWDVIKKM---PMMPNEVVLGSLL 688
Query: 342 AACAQLGDLSVGKSSHAY---VFRNGLERLDNISNAIIDMYMKCGKREAACKVFDSMSNK 398
AAC GD+ + + Y ++ G SN +Y GK + A KV M +
Sbjct: 689 AACRTQGDVELAEKVMKYQVELYPGGDSNYVLFSN----IYAAVGKWDGASKVRREMKER 856
Score = 101 bits (252), Expect = 1e-21
Identities = 72/284 (25%), Positives = 130/284 (45%), Gaps = 1/284 (0%)
Frame = +2
Query: 282 GDMYAVREIFDEFSDKNLVMYNTIMSNYVQHGLAGEVLVVLDEMLQKGQRPDKVTMLSTI 341
GD+ ++FD+ KN+V + ++ +V+ E L EM G PD VT+++ I
Sbjct: 2 GDVDDALKLFDKLPVKNVVSWTVVIGGFVKKECYEEALECFREMQLAGVVPDFVTVIAII 181
Query: 342 AACAQLGDLSVGKSSHAYVFRNGLERLDNISNAIIDMYMKCGKREAACKVFDSMSNKTVV 401
+ACA LG L +G H V + + N++IDMY +CG C
Sbjct: 182 SACANLGALGLGLWVHRLVMKKEFRDNVKVLNSLIDMYARCG-----C------------ 310
Query: 402 TWNSLIAGLVRDGELELALRIFGEMPESNLVSWNTMIGAMVQASMFEEAIDLLREMQNQG 461
+ELA ++F M + NLVSWN++I + ++A+ R M+ +G
Sbjct: 311 --------------IELARQVFDGMSQRNLVSWNSIIVGFAVNGLADKALSFFRSMKKEG 448
Query: 462 IKGDRVTMVGIASACGYLGALDLAKWIYTYIEKNDIHID-MQLGTALVDMFSRCGDPLNA 520
++ + V+ +AC + G +D I+ I+++ + ++ LVD++SR G A
Sbjct: 449 LEPNGVSYTSALTACSHAGLIDEGLKIFADIKRDHRNSPRIEHYGCLVDLYSRAGRLKEA 628
Query: 521 MRVFENMEKRDVSAWTAAIRVKAVEGNAKGAIELFDEMLKQDVK 564
V + M ++ +G +EL ++++K V+
Sbjct: 629 WDVIKKMPMMPNEVVLGSLLAAC---RTQGDVELAEKVMKYQVE 751
>TC80535 weakly similar to GP|14335136|gb|AAK59848.1 At2g15690/F9O13.24
{Arabidopsis thaliana}, partial (55%)
Length = 1727
Score = 199 bits (507), Expect = 3e-51
Identities = 111/383 (28%), Positives = 197/383 (50%)
Frame = +2
Query: 456 EMQNQGIKGDRVTMVGIASACGYLGALDLAKWIYTYIEKNDIHIDMQLGTALVDMFSRCG 515
E+ +GIK D + CG +++ AK ++ Y ++ D ++ +++M+ C
Sbjct: 251 ELMEKGIKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFRSDFKMHNKVIEMYGNCK 430
Query: 516 DPLNAMRVFENMEKRDVSAWTAAIRVKAVEGNAKGAIELFDEMLKQDVKADDFVFVALLT 575
+A RVF++M R++ +W IR A ++LF++M + ++ +A+L+
Sbjct: 431 SMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNELGLEITSETMLAVLS 610
Query: 576 AFSHGGYVDQGRQLFWAMEKIHGVSPQIVHYGCMVDLLGRAGLLEEAFDLMKSMPIKPND 635
A V+ +M+ +G+ P + HY ++D+LG++G L+EA + ++ +P +P
Sbjct: 611 ACGSAEAVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQSGYLKEAEEFIEQLPFEPTV 790
Query: 636 VIWGSFLAACRKHKNVEFANYADEKITQLAPEKVGIHVLLSNIYASAGKWNDVARVRLQM 695
++ + R H +V+ ++ +E I L P K V ++
Sbjct: 791 TVFETLKNYARIHGDVDLEDHVEELIVSLDPSKA---------------------VANKI 907
Query: 696 KEKGFQKVAGSSSIEVHGLIREFTSGDESHTENAQIGLMLQEINCRISQVGYVPDTTNVL 755
+K S ++ I E+ + + I + + GYVPDT VL
Sbjct: 908 PTPPPKKYTAISMLDGKNRIIEYKNPTLYKDDEKLIAMN------SMKDAGYVPDTRYVL 1069
Query: 756 VDVDEQEKEHLLSRHSEKLAMAYGLINTGKGIPIRVVKNLRMCSDCHSFAKLVSKLYGRE 815
D+D++ KE L HSE+LA+AYGLI+T P+R++KNLR+C DCH+ K++S++ GRE
Sbjct: 1070HDIDQEAKEQALLYHSERLAIAYGLISTPPRTPLRIIKNLRVCGDCHNAIKIMSRIVGRE 1249
Query: 816 ITVRDNNRYHFFKEGFCSCRDFW 838
+ VRDN R+H FK+G CSC D+W
Sbjct: 1250LIVRDNKRFHHFKDGKCSCGDYW 1318
Score = 79.0 bits (193), Expect = 7e-15
Identities = 56/210 (26%), Positives = 92/210 (43%), Gaps = 2/210 (0%)
Frame = +2
Query: 106 YAASGLCKEAIFIYLHMIIVMGIVPDNFTFPFLLSACSKIMAFSEGVQVHGVVVKMGLVK 165
+ G KEA+ ++ GI D F L C K + + +VH ++
Sbjct: 218 FCQEGKVKEAL-----ELMEKGIKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFRS 382
Query: 166 DLFVANSLIHFYAACGKVDLGRKVFDEMLERNVVSWTSLINGYSVVNMAKEAVCLFFEMV 225
D + N +I Y C + R+VFD M RN+ SW +I GY+ M E + LF +M
Sbjct: 383 DFKMHNKVIEMYGNCKSMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMN 562
Query: 226 EVGVEPNPVTMVCAISACAKLKDLELGKKVCNLM-TELGVKSNTLVVNALLDMYMKCGDM 284
E+G+E TM+ +SAC + +E M ++ G++ LLD+ + G +
Sbjct: 563 ELGLEITSETMLAVLSACGSAEAVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQSGYL 742
Query: 285 YAVREIFDEFSDKNLVMYNTIMSNYVQ-HG 313
E ++ + V + NY + HG
Sbjct: 743 KEAEEFIEQLPFEPTVTVFETLKNYARIHG 832
Score = 58.2 bits (139), Expect = 1e-08
Identities = 30/122 (24%), Positives = 60/122 (48%)
Frame = +2
Query: 223 EMVEVGVEPNPVTMVCAISACAKLKDLELGKKVCNLMTELGVKSNTLVVNALLDMYMKCG 282
E++E G++ + C K K +E KKV + + +S+ + N +++MY C
Sbjct: 251 ELMEKGIKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTFRSDFKMHNKVIEMYGNCK 430
Query: 283 DMYAVREIFDEFSDKNLVMYNTIMSNYVQHGLAGEVLVVLDEMLQKGQRPDKVTMLSTIA 342
M R +FD ++N+ ++ ++ Y + E L + ++M + G TML+ ++
Sbjct: 431 SMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQMNELGLEITSETMLAVLS 610
Query: 343 AC 344
AC
Sbjct: 611 AC 616
Score = 55.8 bits (133), Expect = 7e-08
Identities = 46/186 (24%), Positives = 81/186 (42%), Gaps = 5/186 (2%)
Frame = +2
Query: 306 MSNYVQHGLAGEVLVVLDEMLQKGQRPDKVTMLSTIAACAQLGDLSVGKSSHAYVFRNGL 365
++ + Q G E L E+++KG + D C + + K H Y ++
Sbjct: 209 LTRFCQEGKVKEAL----ELMEKGIKADANCFEILFDLCGKSKSVEDAKKVHDYFLQSTF 376
Query: 366 ERLDNISNAIIDMYMKCGKREAACKVFDSMSNKTVVTWNSLIAGLVRDGELELALRIFGE 425
+ N +I+MY C A +VFD M N+ + +W+ +I G + L++F +
Sbjct: 377 RSDFKMHNKVIEMYGNCKSMTDARRVFDHMPNRNMDSWHMMIRGYANSTMGDEGLQLFEQ 556
Query: 426 MPESNL-VSWNTM---IGAMVQASMFEEAIDLLREMQNQ-GIKGDRVTMVGIASACGYLG 480
M E L ++ TM + A A E+A L M+++ GI+ +G+ G G
Sbjct: 557 MNELGLEITSETMLAVLSACGSAEAVEDAYIYLESMKSKYGIEPGVEHYMGLLDVLGQSG 736
Query: 481 ALDLAK 486
L A+
Sbjct: 737 YLKEAE 754
>BG455368 weakly similar to GP|9759524|dbj selenium-binding protein-like
{Arabidopsis thaliana}, partial (33%)
Length = 687
Score = 191 bits (484), Expect = 1e-48
Identities = 96/221 (43%), Positives = 138/221 (62%), Gaps = 1/221 (0%)
Frame = +1
Query: 619 LEEAFDLMKSMPIKPNDVIWGSFLAACRKHKNVEFANYADEKITQLAPEKVGIHVLLSNI 678
+E+A L+++MP++PN +WG+ L A H N + A A + +L P+ +G ++LLS
Sbjct: 7 VEKALQLVQTMPMEPNGGVWGALLGASHIHGNPDVAEIASRSLFELEPDNLGNYLLLSKT 186
Query: 679 YASAGKWNDVARVRLQMKEKGFQKVAGSSSIEV-HGLIREFTSGDESHTENAQIGLMLQE 737
YA A KW+DV+RVR M+EK +K G S +E +G+I EF +GD H E +I L +
Sbjct: 187 YALAAKWDDVSRVRKLMREKQLRKNPGCSWVEAKNGIIHEFFAGDVKHPEINEIKKALDD 366
Query: 738 INCRISQVGYVPDTTNVLVDVDEQEKEHLLSRHSEKLAMAYGLINTGKGIPIRVVKNLRM 797
+ R+ GY P +V D+D++ K LL HSEKLA+AYGL++T G I+++KNLR+
Sbjct: 367 LLQRLKCTGYQPKLNSVPYDIDDEGKRCLLVSHSEKLALAYGLLSTDAGSTIKIMKNLRI 546
Query: 798 CSDCHSFAKLVSKLYGREITVRDNNRYHFFKEGFCSCRDFW 838
C DCH S L GR+I VRDN +H F G CSC +FW
Sbjct: 547 CEDCHIVMCGASXLTGRKIIVRDNMXFHHFLNGACSCNNFW 669
>TC90451 similar to PIR|H84508|H84508 hypothetical protein At2g13600
[imported] - Arabidopsis thaliana, partial (12%)
Length = 1362
Score = 139 bits (350), Expect(2) = 3e-36
Identities = 76/234 (32%), Positives = 131/234 (55%), Gaps = 4/234 (1%)
Frame = +2
Query: 194 LERNVVS--WTSLINGYSVVNMAKEAVCLFFEMVEVGVEPNPVTMVCAISACAKLKDLEL 251
LE N S W ++I Y+ + + A+ ++ M+ GV P+ T+ + A ++ ++L
Sbjct: 362 LESNPASFNWNNIIRSYTRLESPQNALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFAIQL 541
Query: 252 GKKVCNLMTELGVKSNTLVVNALLDMYMKCGDMYAVREIFDEFSDKNLVMYNTIMSNYVQ 311
G++V + +LG++SN + +++Y K GD + ++FDE + L +N ++S Q
Sbjct: 542 GQQVHSYGIKLGLQSNEYCESGFINLYCKAGDFDSAHKVFDENHEPKLGSWNALISGLSQ 721
Query: 312 HGLAGEVLVVLDEMLQKGQRPDKVTMLSTIAACAQLGDLSVGKSSHAYVFRNGLERLDNI 371
GLA + +VV +M + G PD +TM+S ++AC +GDL + H YVF+ I
Sbjct: 722 GGLAMDAIVVFVDMKRHGFEPDGITMVSVMSACGSIGDLYLALQLHKYVFQAKTNEWTVI 901
Query: 372 --SNAIIDMYMKCGKREAACKVFDSMSNKTVVTWNSLIAGLVRDGELELALRIF 423
SN++IDMY KCG+ + A +VF +M ++ V +W S+I G G + AL F
Sbjct: 902 LMSNSLIDMYGKCGRMDLAYEVFATMEDRNVSSWTSMIVGYAMHGHAKEALGCF 1063
Score = 138 bits (347), Expect = 1e-32
Identities = 76/225 (33%), Positives = 121/225 (53%), Gaps = 2/225 (0%)
Frame = +2
Query: 97 YTCNTLIRGYAASGLCKEAIFIYLHMIIVMGIVPDNFTFPFLLSACSKIMAFSEGVQVHG 156
+ N +IR Y + A+ IY+ M+ G++PD +T P +L A S+ A G QVH
Sbjct: 383 FNWNNIIRSYTRLESPQNALRIYVSMLRA-GVLPDRYTLPIVLKAVSQSFAIQLGQQVHS 559
Query: 157 VVVKMGLVKDLFVANSLIHFYAACGKVDLGRKVFDEMLERNVVSWTSLINGYSVVNMAKE 216
+K+GL + + + I+ Y G D KVFDE E + SW +LI+G S +A +
Sbjct: 560 YGIKLGLQSNEYCESGFINLYCKAGDFDSAHKVFDENHEPKLGSWNALISGLSQGGLAMD 739
Query: 217 AVCLFFEMVEVGVEPNPVTMVCAISACAKLKDLELGKKVCNLMTELGVKSNTLVV--NAL 274
A+ +F +M G EP+ +TMV +SAC + DL L ++ + + T+++ N+L
Sbjct: 740 AIVVFVDMKRHGFEPDGITMVSVMSACGSIGDLYLALQLHKYVFQAKTNEWTVILMSNSL 919
Query: 275 LDMYMKCGDMYAVREIFDEFSDKNLVMYNTIMSNYVQHGLAGEVL 319
+DMY KCG M E+F D+N+ + +++ Y HG A E L
Sbjct: 920 IDMYGKCGRMDLAYEVFATMEDRNVSSWTSMIVGYAMHGHAKEAL 1054
Score = 112 bits (280), Expect(3) = 1e-46
Identities = 75/256 (29%), Positives = 127/256 (49%), Gaps = 2/256 (0%)
Frame = +2
Query: 302 YNTIMSNYVQHGLAGEVLVVLDEMLQKGQRPDKVTMLSTIAACAQLGDLSVGKSSHAYVF 361
+N I+ +Y + L + ML+ G PD+ T+ + A +Q + +G+ H+Y
Sbjct: 389 WNNIIRSYTRLESPQNALRIYVSMLRAGVLPDRYTLPIVLKAVSQSFAIQLGQQVHSYGI 568
Query: 362 RNGLERLDNISNAIIDMYMKCGKREAACKVFDSMSNKTVVTWNSLIAGLVRDGELELALR 421
+ GL+ + + I++Y K G ++A KVFD + +WN+LI+GL + G LA+
Sbjct: 569 KLGLQSNEYCESGFINLYCKAGDFDSAHKVFDENHEPKLGSWNALISGLSQGG---LAM- 736
Query: 422 IFGEMPESNLVSWNTMIGAMVQASMFEEAIDLLREMQNQGIKGDRVTMVGIASACGYLGA 481
+AI + +M+ G + D +TMV + SACG +G
Sbjct: 737 ---------------------------DAIVVFVDMKRHGFEPDGITMVSVMSACGSIGD 835
Query: 482 LDLAKWIYTYI--EKNDIHIDMQLGTALVDMFSRCGDPLNAMRVFENMEKRDVSAWTAAI 539
L LA ++ Y+ K + + + +L+DM+ +CG A VF ME R+VS+WT+ I
Sbjct: 836 LYLALQLHKYVFQAKTNEWTVILMSNSLIDMYGKCGRMDLAYEVFATMEDRNVSSWTSMI 1015
Query: 540 RVKAVEGNAKGAIELF 555
A+ G+AK A+ F
Sbjct: 1016VGYAMHGHAKEALGCF 1063
Score = 93.2 bits (230), Expect(3) = 1e-46
Identities = 41/79 (51%), Positives = 58/79 (72%)
Frame = +1
Query: 563 VKADDFVFVALLTAFSHGGYVDQGRQLFWAMEKIHGVSPQIVHYGCMVDLLGRAGLLEEA 622
VK + F+ +L+A HGG V +GR F M+ I+G++PQ+ HYGCMVDLLGRAGL ++A
Sbjct: 1087 VKPNYVTFIGVLSACVHGGTVQEGRFYFDMMKNIYGITPQLQHYGCMVDLLGRAGLFDDA 1266
Query: 623 FDLMKSMPIKPNDVIWGSF 641
+++ MP+KPN V+WG F
Sbjct: 1267 RRMVEEMPMKPNSVVWGVF 1323
Score = 85.1 bits (209), Expect(2) = 8e-19
Identities = 55/143 (38%), Positives = 77/143 (53%), Gaps = 2/143 (1%)
Frame = +2
Query: 89 DEGTKCSLYTCNTLIRGYAASGLCKEAIFIYLHMIIVMGIVPDNFTFPFLLSACSKIMAF 148
DE + L + N LI G + GL +AI +++ M G PD T ++SAC I
Sbjct: 662 DENHEPKLGSWNALISGLSQGGLAMDAIVVFVDMKR-HGFEPDGITMVSVMSACGSIGDL 838
Query: 149 SEGVQVHGVV--VKMGLVKDLFVANSLIHFYAACGKVDLGRKVFDEMLERNVVSWTSLIN 206
+Q+H V K + ++NSLI Y CG++DL +VF M +RNV SWTS+I
Sbjct: 839 YLALQLHKYVFQAKTNEWTVILMSNSLIDMYGKCGRMDLAYEVFATMEDRNVSSWTSMIV 1018
Query: 207 GYSVVNMAKEAVCLFFEMVEVGV 229
GY++ AKEA+ F M VGV
Sbjct: 1019GYAMHGHAKEALGCFHCMKGVGV 1087
Score = 54.3 bits (129), Expect = 2e-07
Identities = 54/269 (20%), Positives = 108/269 (40%), Gaps = 10/269 (3%)
Frame = +2
Query: 396 SNKTVVTWNSLIAGLVRDGELELALRIFGEMPESNLVSWNTMIGAMVQASMFEEAIDLLR 455
SN WN++I R + ALRI+ + +LR
Sbjct: 368 SNPASFNWNNIIRSYTRLESPQNALRIY---------------------------VSMLR 466
Query: 456 EMQNQGIKGDRVTMVGIASACGYLGALDLAKWIYTYIEKNDIHIDMQLGTALVDMFSRCG 515
G+ DR T+ + A A+ L + +++Y K + + + ++++ + G
Sbjct: 467 ----AGVLPDRYTLPIVLKAVSQSFAIQLGQQVHSYGIKLGLQSNEYCESGFINLYCKAG 634
Query: 516 DPLNAMRVFENMEKRDVSAWTAAIRVKAVEGNAKGAIELFDEMLKQDVKADDFVFVALLT 575
D +A +VF+ + + +W A I + G A AI +F +M + + D V++++
Sbjct: 635 DFDSAHKVFDENHEPKLGSWNALISGLSQGGLAMDAIVVFVDMKRHGFEPDGITMVSVMS 814
Query: 576 AFSHGG----------YVDQGRQLFWAMEKIHGVSPQIVHYGCMVDLLGRAGLLEEAFDL 625
A G YV Q + W + I+ ++D+ G+ G ++ A+++
Sbjct: 815 ACGSIGDLYLALQLHKYVFQAKTNEWTV---------ILMSNSLIDMYGKCGRMDLAYEV 967
Query: 626 MKSMPIKPNDVIWGSFLAACRKHKNVEFA 654
+M + N W S + H + + A
Sbjct: 968 FATMEDR-NVSSWTSMIVGYAMHGHAKEA 1051
Score = 32.0 bits (71), Expect(2) = 3e-36
Identities = 23/71 (32%), Positives = 38/71 (53%), Gaps = 2/71 (2%)
Frame = +1
Query: 459 NQGIKGDRVTMVGIASACGYLGALDLAKWIYTYIEKNDIHIDMQLG--TALVDMFSRCGD 516
++G+K + VT +G+ SAC + G + ++ Y + KN I QL +VD+ R G
Sbjct: 1078 SRGVKPNYVTFIGVLSACVHGGTVQEGRF-YFDMMKNIYGITPQLQHYGCMVDLLGRAGL 1254
Query: 517 PLNAMRVFENM 527
+A R+ E M
Sbjct: 1255 FDDARRMVEEM 1287
Score = 27.3 bits (59), Expect(2) = 8e-19
Identities = 17/71 (23%), Positives = 33/71 (45%), Gaps = 1/71 (1%)
Frame = +1
Query: 228 GVEPNPVTMVCAISACAKLKDLELGKKVCNLMTEL-GVKSNTLVVNALLDMYMKCGDMYA 286
GV+PN VT + +SAC ++ G+ ++M + G+ ++D+ + G
Sbjct: 1084 GVKPNYVTFIGVLSACVHGGTVQEGRFYFDMMKNIYGITPQLQHYGCMVDLLGRAGLFDD 1263
Query: 287 VREIFDEFSDK 297
R + +E K
Sbjct: 1264 ARRMVEEMPMK 1296
Score = 21.2 bits (43), Expect(3) = 1e-46
Identities = 7/14 (50%), Positives = 9/14 (64%)
Frame = +2
Query: 639 GSFLAACRKHKNVE 652
G + AC KH NV+
Sbjct: 1316 GCLMGACEKHGNVD 1357
>AW776322 weakly similar to PIR|H84859|H84 hypothetical protein At2g42920
[imported] - Arabidopsis thaliana, partial (27%)
Length = 633
Score = 162 bits (411), Expect = 4e-40
Identities = 81/208 (38%), Positives = 128/208 (60%), Gaps = 1/208 (0%)
Frame = +1
Query: 525 ENMEKRDVSAWTAAIRVKAVEGNAKGAIELFDEMLKQDV-KADDFVFVALLTAFSHGGYV 583
E +R +S W + I A+ G+ + A E F ++ + K D F+ +LTA H G +
Sbjct: 1 ETCPRRGLSCWNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAI 180
Query: 584 DQGRQLFWAMEKIHGVSPQIVHYGCMVDLLGRAGLLEEAFDLMKSMPIKPNDVIWGSFLA 643
++ R F M + + P I HY C+VD+LG+AGLLEEA +L+K MP+KP+ +IWGS L+
Sbjct: 181 NKARDYFELMMNKYEIEPSIKHYTCIVDVLGQAGLLEEAEELIKGMPLKPDAIIWGSLLS 360
Query: 644 ACRKHKNVEFANYADEKITQLAPEKVGIHVLLSNIYASAGKWNDVARVRLQMKEKGFQKV 703
+CRKH+NV+ A A +++ +L P +VL+SN++A++ K+ + RL MKE +K
Sbjct: 361 SCRKHRNVQIARRAAQRVYELNPSDASGYVLMSNVHAASNKFEEAIEQRLLMKENLTEKE 540
Query: 704 AGSSSIEVHGLIREFTSGDESHTENAQI 731
G SSIE++G + EF +G H + +I
Sbjct: 541 PGCSSIELYGEVHEFIAGGRLHPKTQEI 624
Score = 43.5 bits (101), Expect = 3e-04
Identities = 28/112 (25%), Positives = 55/112 (49%), Gaps = 3/112 (2%)
Frame = +1
Query: 427 PESNLVSWNTMIGAMVQASMFEEAIDLLREMQNQGI-KGDRVTMVGIASACGYLGALDLA 485
P L WN++I + EA + ++++ + K D V+ +G+ +AC +LGA++ A
Sbjct: 10 PRRGLSCWNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAINKA 189
Query: 486 K-WIYTYIEKNDIHIDMQLGTALVDMFSRCGDPLNAMRVFENME-KRDVSAW 535
+ + + K +I ++ T +VD+ + G A + + M K D W
Sbjct: 190 RDYFELMMNKYEIEPSIKHYTCIVDVLGQAGLLEEAEELIKGMPLKPDAIIW 345
Score = 37.7 bits (86), Expect = 0.018
Identities = 36/163 (22%), Positives = 72/163 (44%), Gaps = 7/163 (4%)
Frame = +1
Query: 297 KNLVMYNTIMSNYVQHGLAGEVLVVLDEM-LQKGQRPDKVTMLSTIAACAQLGDLSVGKS 355
+ L +N+I+ +G E ++ K +PD V+ + + AC LG ++ +
Sbjct: 16 RGLSCWNSIIIGLAMNGHEREAFEFFSKLESSKLLKPDSVSFIGVLTACKHLGAINKAR- 192
Query: 356 SHAYVFRNGLERLDNISN--AIIDMYMKCGKREAACKVFDSMSNK-TVVTWNSLIAGLVR 412
+ + N E +I + I+D+ + G E A ++ M K + W SL++ +
Sbjct: 193 DYFELMMNKYEIEPSIKHYTCIVDVLGQAGLLEEAEELIKGMPLKPDAIIWGSLLSSCRK 372
Query: 413 DGELELALRI---FGEMPESNLVSWNTMIGAMVQASMFEEAID 452
+++A R E+ S+ + M ++ FEEAI+
Sbjct: 373 HRNVQIARRAAQRVYELNPSDASGYVLMSNVHAASNKFEEAIE 501
>BE941714 weakly similar to GP|11994734|dbj selenium-binding protein-like
{Arabidopsis thaliana}, partial (10%)
Length = 619
Score = 158 bits (399), Expect = 9e-39
Identities = 84/205 (40%), Positives = 126/205 (60%), Gaps = 5/205 (2%)
Frame = +2
Query: 497 IHIDMQLGTALVDMFSRCGDPLNAMRVFENMEKRDVSAWTAAIRVKAVEGNAKGAIELFD 556
+ + ++L T+L+DM+++CG+ A R+F++M RDV W A I A+ G+ KGA++LF
Sbjct: 2 VPLSVRLSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFY 181
Query: 557 EMLKQDVKADDFVFVALLTAFSHGGYVDQGRQLFWAMEKIHGVSPQIVHYGCMVDLLGRA 616
+M K VK DD F+A+ TA S+ G +G L M ++ + P+ HYGC+VDLL RA
Sbjct: 182 DMEKVGVKPDDITFIAVFTACSYSGMAYEGLMLLDKMCSVYNIVPKSEHYGCLVDLLSRA 361
Query: 617 GLLEEAFDLMKSMPIKPN----DVIWGSFLAACRKHKNVEFANYADEKITQLAPE-KVGI 671
GL EEA +++ + N + W +FL+AC H + A A EK+ QL G+
Sbjct: 362 GLFEEAMVMIRKITNSWNGSEETLAWRAFLSACCNHGETQLAELAAEKVLQLDNHIHSGV 541
Query: 672 HVLLSNIYASAGKWNDVARVRLQMK 696
+VLLSN+YA++GK ND RV+ MK
Sbjct: 542 YVLLSNLYAASGKHNDARRVKDMMK 616
Score = 73.6 bits (179), Expect = 3e-13
Identities = 47/176 (26%), Positives = 92/176 (51%), Gaps = 15/176 (8%)
Frame = +2
Query: 169 VANSLIHFYAACGKVDLGRKVFDEMLERNVVSWTSLINGYSVVNMAKEAVCLFFEMVEVG 228
++ SL+ YA CG ++L +++FD M R+VV W ++I+G ++ K A+ LF++M +VG
Sbjct: 20 LSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDMEKVG 199
Query: 229 VEPNPVTMVCAISACA----KLKDLELGKKVCNLMTELGVKSNTLVVNALLDMYMKCG-- 282
V+P+ +T + +AC+ + L L K+C++ + + L+D+ + G
Sbjct: 200 VKPDDITFIAVFTACSYSGMAYEGLMLLDKMCSV---YNIVPKSEHYGCLVDLLSRAGLF 370
Query: 283 --DMYAVREIFDEFS-DKNLVMYNTIMSNYVQHG------LAGEVLVVLDEMLQKG 329
M +R+I + ++ + + + +S HG LA E ++ LD + G
Sbjct: 371 EEAMVMIRKITNSWNGSEETLAWRAFLSACCNHGETQLAELAAEKVLQLDNHIHSG 538
Score = 72.0 bits (175), Expect = 9e-13
Identities = 56/231 (24%), Positives = 100/231 (43%), Gaps = 12/231 (5%)
Frame = +2
Query: 371 ISNAIIDMYMKCGKREAACKVFDSMSNKTVVTWNSLIAGLVRDGELELALRIFGEMPESN 430
+S +++DMY KCG E A ++FDSM+ + VV WN++I+G+ G+ + AL++F
Sbjct: 20 LSTSLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLF------- 178
Query: 431 LVSWNTMIGAMVQASMFEEAIDLLREMQNQGIKGDRVTMVGIASACGYLG-------ALD 483
+M+ G+K D +T + + +AC Y G LD
Sbjct: 179 ------------------------YDMEKVGVKPDDITFIAVFTACSYSGMAYEGLMLLD 286
Query: 484 LAKWIYTYIEKNDIHIDMQLGTALVDMFSRCGDPLNAMRVFENME-----KRDVSAWTAA 538
+Y + K++ + LVD+ SR G AM + + + AW A
Sbjct: 287 KMCSVYNIVPKSEHY------GCLVDLLSRAGLFEEAMVMIRKITNSWNGSEETLAWRAF 448
Query: 539 IRVKAVEGNAKGAIELFDEMLKQDVKADDFVFVALLTAFSHGGYVDQGRQL 589
+ G + A +++L+ D V+V L ++ G + R++
Sbjct: 449 LSACCNHGETQLAELAAEKVLQLDNHIHSGVYVLLSNLYAASGKHNDARRV 601
Score = 62.4 bits (150), Expect = 7e-10
Identities = 38/161 (23%), Positives = 76/161 (46%), Gaps = 14/161 (8%)
Frame = +2
Query: 273 ALLDMYMKCGDMYAVREIFDEFSDKNLVMYNTIMSNYVQHGLAGEVLVVLDEMLQKGQRP 332
+LLDMY KCG++ + +FD + +++V +N ++S HG L + +M + G +P
Sbjct: 29 SLLDMYAKCGNLELAKRLFDSMNMRDVVCWNAMISGMAMHGDGKGALKLFYDMEKVGVKP 208
Query: 333 DKVTMLSTIAACAQLGDLSVGKSSHAYVFRNGLERLDNISN---------AIIDMYMKCG 383
D +T ++ AC+ S AY L+++ ++ N ++D+ + G
Sbjct: 209 DDITFIAVFTACSY--------SGMAYEGLMLLDKMCSVYNIVPKSEHYGCLVDLLSRAG 364
Query: 384 KREAACKVFDSMSN-----KTVVTWNSLIAGLVRDGELELA 419
E A + ++N + + W + ++ GE +LA
Sbjct: 365 LFEEAMVMIRKITNSWNGSEETLAWRAFLSACCNHGETQLA 487
Score = 29.3 bits (64), Expect = 6.6
Identities = 19/55 (34%), Positives = 29/55 (52%)
Frame = +2
Query: 100 NTLIRGYAASGLCKEAIFIYLHMIIVMGIVPDNFTFPFLLSACSKIMAFSEGVQV 154
N +I G A G K A+ ++ M V G+ PD+ TF + +ACS EG+ +
Sbjct: 119 NAMISGMAMHGDGKGALKLFYDMEKV-GVKPDDITFIAVFTACSYSGMAYEGLML 280
>CA921534 similar to PIR|D96516|D96 F16N3.14 [imported] - Arabidopsis
thaliana, partial (18%)
Length = 790
Score = 148 bits (374), Expect = 7e-36
Identities = 86/230 (37%), Positives = 131/230 (56%)
Frame = -1
Query: 609 MVDLLGRAGLLEEAFDLMKSMPIKPNDVIWGSFLAACRKHKNVEFANYADEKITQLAPEK 668
+V++ G A L +A + M++MPI+P +W + R H ++E + ADE +T L P K
Sbjct: 769 VVNIFGCAVGLMKAHEFMRNMPIEPGVELWETLRNFARIHGDLEREDCADELLTVLDPSK 590
Query: 669 VGIHVLLSNIYASAGKWNDVARVRLQMKEKGFQKVAGSSSIEVHGLIREFTSGDESHTEN 728
A+A K V L ++K + + +E + E+ E
Sbjct: 589 -----------AAADK------VPLPQRKKQ----SAINMLEEKNRVSEYRCNMPYKEEG 473
Query: 729 AQIGLMLQEINCRISQVGYVPDTTNVLVDVDEQEKEHLLSRHSEKLAMAYGLINTGKGIP 788
+ L+ + ++ + GYVPDT VL D+DE+EKE L HSE+LA+AYGLI+T
Sbjct: 472 ---DVKLRGLTGQMREAGYVPDTRYVLHDIDEEEKEKALQYHSERLAIAYGLISTPPRTT 302
Query: 789 IRVVKNLRMCSDCHSFAKLVSKLYGREITVRDNNRYHFFKEGFCSCRDFW 838
+R++KNLR+C DCH+ K++SK+ GRE+ VRDN R+H FK+G CSC D+W
Sbjct: 301 LRIIKNLRICGDCHNAIKIMSKIVGRELIVRDNKRFHHFKDGKCSCGDYW 152
>BF633384 similar to PIR|T46179|T46 hypothetical protein T8H10.30 -
Arabidopsis thaliana, partial (22%)
Length = 595
Score = 142 bits (358), Expect = 5e-34
Identities = 72/196 (36%), Positives = 111/196 (55%), Gaps = 6/196 (3%)
Frame = +1
Query: 535 WTAAIRVKAVEGNAKGAIELFDEMLKQ-----DVKADDFVFVALLTAFSHGGYVDQGRQL 589
W I + G + A++LF M+++ +++ ++ ++A+ + SH G VD+G L
Sbjct: 7 WNVLIMAYGMHGKGEEALKLFRRMVEEGDNNREIRPNEVTYIAIFASLSHSGMVDEGLNL 186
Query: 590 FWAMEKIHGVSPQIVHYGCMVDLLGRAGLLEEAFDLMKSMPIKPNDV-IWGSFLAACRKH 648
F+ M+ HG+ P HY C+VDLLGR+G +EEA++L+K+MP V W S L AC+ H
Sbjct: 187 FYTMKAKHGIEPTSDHYACLVDLLGRSGQIEEAYNLIKTMPSNMKKVDAWSSLLGACKIH 366
Query: 649 KNVEFANYADEKITQLAPEKVGIHVLLSNIYASAGKWNDVARVRLQMKEKGFQKVAGSSS 708
+N+E A + + L P +VLLSNIY+SAG W+ VR +MKEKG +K
Sbjct: 367 QNLEIGEIAAKNLFVLDPNVASYYVLLSNIYSSAGLWDQAIDVRKKMKEKGVRKNLDVVG 546
Query: 709 IEVHGLIREFTSGDES 724
+ + F GD S
Sbjct: 547 LNMAMRFTSFWQGDVS 594
Score = 42.7 bits (99), Expect = 6e-04
Identities = 39/162 (24%), Positives = 65/162 (40%), Gaps = 8/162 (4%)
Frame = +1
Query: 432 VSWNTMIGAMVQASMFEEAIDLLREMQNQG-----IKGDRVTMVGIASACGYLGALDLA- 485
++WN +I A EEA+ L R M +G I+ + VT + I ++ + G +D
Sbjct: 1 ITWNVLIMAYGMHGKGEEALKLFRRMVEEGDNNREIRPNEVTYIAIFASLSHSGMVDEGL 180
Query: 486 KWIYTYIEKNDIHIDMQLGTALVDMFSRCGDPLNAMRVFENM--EKRDVSAWTAAIRVKA 543
YT K+ I LVD+ R G A + + M + V AW++ +
Sbjct: 181 NLFYTMKAKHGIEPTSDHYACLVDLLGRSGQIEEAYNLIKTMPSNMKKVDAWSSLLGACK 360
Query: 544 VEGNAKGAIELFDEMLKQDVKADDFVFVALLTAFSHGGYVDQ 585
+ N + + D + +V L +S G DQ
Sbjct: 361 IHQNLEIGEIAAKNLFVLDPNVASY-YVLLSNIYSSAGLWDQ 483
Score = 30.4 bits (67), Expect = 2.9
Identities = 16/49 (32%), Positives = 25/49 (50%), Gaps = 5/49 (10%)
Frame = +1
Query: 199 VSWTSLINGYSVVNMAKEAVCLFFEMVEVG-----VEPNPVTMVCAISA 242
++W LI Y + +EA+ LF MVE G + PN VT + ++
Sbjct: 1 ITWNVLIMAYGMHGKGEEALKLFRRMVEEGDNNREIRPNEVTYIAIFAS 147
>BE202961 weakly similar to GP|6016735|gb|A hypothetical protein {Arabidopsis
thaliana}, partial (14%)
Length = 613
Score = 135 bits (340), Expect = 6e-32
Identities = 72/199 (36%), Positives = 110/199 (55%)
Frame = +2
Query: 223 EMVEVGVEPNPVTMVCAISACAKLKDLELGKKVCNLMTELGVKSNTLVVNALLDMYMKCG 282
EM++ ++PN VT+V + ACA + +LE G K+ L G + T V AL+DMYMKC
Sbjct: 17 EMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDMYMKCF 196
Query: 283 DMYAVREIFDEFSDKNLVMYNTIMSNYVQHGLAGEVLVVLDEMLQKGQRPDKVTMLSTIA 342
+IF+ K+++ + + S Y +G+ E + V ML G RPD + ++ +
Sbjct: 197 SPEKAVDIFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIALVKILT 376
Query: 343 ACAQLGDLSVGKSSHAYVFRNGLERLDNISNAIIDMYMKCGKREAACKVFDSMSNKTVVT 402
++LG L HA+V +NG E I ++I++Y KC E A KVF M+ K VVT
Sbjct: 377 TVSELGILQQAVCFHAFVIKNGFENNQFIGASLIEVYAKCSSIEDANKVFKGMTYKDVVT 556
Query: 403 WNSLIAGLVRDGELELALR 421
W+S+IA G+ E AL+
Sbjct: 557 WSSIIAAYGFHGQGEEALK 613
Score = 101 bits (252), Expect = 1e-21
Identities = 58/230 (25%), Positives = 104/230 (45%)
Frame = +2
Query: 324 EMLQKGQRPDKVTMLSTIAACAQLGDLSVGKSSHAYVFRNGLERLDNISNAIIDMYMKCG 383
EML K +P+ VT++S + ACA + +L G H G E +S A++DMYMKC
Sbjct: 17 EMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDMYMKCF 196
Query: 384 KREAACKVFDSMSNKTVVTWNSLIAGLVRDGELELALRIFGEMPESNLVSWNTMIGAMVQ 443
E A + IF MP+ ++++W +
Sbjct: 197 SPEKA-------------------------------VDIFNRMPKKDVIAWAVLFSGYAD 283
Query: 444 ASMFEEAIDLLREMQNQGIKGDRVTMVGIASACGYLGALDLAKWIYTYIEKNDIHIDMQL 503
M E++ + R M + G + D + +V I + LG L A + ++ KN + +
Sbjct: 284 NGMVHESMWVFRNMLSSGTRPDAIALVKILTTVSELGILQQAVCFHAFVIKNGFENNQFI 463
Query: 504 GTALVDMFSRCGDPLNAMRVFENMEKRDVSAWTAAIRVKAVEGNAKGAIE 553
G +L++++++C +A +VF+ M +DV W++ I G + A++
Sbjct: 464 GASLIEVYAKCSSIEDANKVFKGMTYKDVVTWSSIIAAYGFHGQGEEALK 613
Score = 89.0 bits (219), Expect = 7e-18
Identities = 47/192 (24%), Positives = 100/192 (51%)
Frame = +2
Query: 128 IVPDNFTFPFLLSACSKIMAFSEGVQVHGVVVKMGLVKDLFVANSLIHFYAACGKVDLGR 187
I P+ T +L AC+ I EG+++H + V G + V+ +L+ Y C +
Sbjct: 35 IKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDMYMKCFSPEKAV 214
Query: 188 KVFDEMLERNVVSWTSLINGYSVVNMAKEAVCLFFEMVEVGVEPNPVTMVCAISACAKLK 247
+F+ M +++V++W L +GY+ M E++ +F M+ G P+ + +V ++ ++L
Sbjct: 215 DIFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIALVKILTTVSELG 394
Query: 248 DLELGKKVCNLMTELGVKSNTLVVNALLDMYMKCGDMYAVREIFDEFSDKNLVMYNTIMS 307
L+ + + G ++N + +L+++Y KC + ++F + K++V +++I++
Sbjct: 395 ILQQAVCFHAFVIKNGFENNQFIGASLIEVYAKCSSIEDANKVFKGMTYKDVVTWSSIIA 574
Query: 308 NYVQHGLAGEVL 319
Y HG E L
Sbjct: 575 AYGFHGQGEEAL 610
Score = 78.6 bits (192), Expect = 9e-15
Identities = 56/204 (27%), Positives = 91/204 (44%)
Frame = +2
Query: 451 IDLLREMQNQGIKGDRVTMVGIASACGYLGALDLAKWIYTYIEKNDIHIDMQLGTALVDM 510
+DL EM ++ IK + VT+V + AC + L+ I+ ++ + TAL+DM
Sbjct: 2 LDLSSEMLDKRIKPNWVTVVSVLRACACISNLEEGMKIHELAVNYGFEMETTVSTALMDM 181
Query: 511 FSRCGDPLNAMRVFENMEKRDVSAWTAAIRVKAVEGNAKGAIELFDEMLKQDVKADDFVF 570
+ +C P A+ +F M K+DV AW A G ++ +F ML + D
Sbjct: 182 YMKCFSPEKAVDIFNRMPKKDVIAWAVLFSGYADNGMVHESMWVFRNMLSSGTRPDAIAL 361
Query: 571 VALLTAFSHGGYVDQGRQLFWAMEKIHGVSPQIVHYGCMVDLLGRAGLLEEAFDLMKSMP 630
V +LT S G + Q F A +G ++++ + +E+A + K M
Sbjct: 362 VKILTTVSELGILQQA-VCFHAFVIKNGFENNQFIGASLIEVYAKCSSIEDANKVFKGMT 538
Query: 631 IKPNDVIWGSFLAACRKHKNVEFA 654
K + V W S +AA H E A
Sbjct: 539 YK-DVVTWSSIIAAYGFHGQGEEA 607
Score = 68.9 bits (167), Expect = 7e-12
Identities = 36/117 (30%), Positives = 62/117 (52%)
Frame = +2
Query: 102 LIRGYAASGLCKEAIFIYLHMIIVMGIVPDNFTFPFLLSACSKIMAFSEGVQVHGVVVKM 161
L GYA +G+ E+++++ +M+ G PD +L+ S++ + V H V+K
Sbjct: 263 LFSGYADNGMVHESMWVFRNMLS-SGTRPDAIALVKILTTVSELGILQQAVCFHAFVIKN 439
Query: 162 GLVKDLFVANSLIHFYAACGKVDLGRKVFDEMLERNVVSWTSLINGYSVVNMAKEAV 218
G + F+ SLI YA C ++ KVF M ++VV+W+S+I Y +EA+
Sbjct: 440 GFENNQFIGASLIEVYAKCSSIEDANKVFKGMTYKDVVTWSSIIAAYGFHGQGEEAL 610
>AL370459 weakly similar to GP|20146256|d hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (11%)
Length = 477
Score = 132 bits (332), Expect = 5e-31
Identities = 63/124 (50%), Positives = 84/124 (66%)
Frame = +1
Query: 715 IREFTSGDESHTENAQIGLMLQEINCRISQVGYVPDTTNVLVDVDEQEKEHLLSRHSEKL 774
+ +F GD H + QI L E+ +I VGYVP+T VL DV++++KE L +HSEKL
Sbjct: 10 VHKFHVGDTLHPKAQQIYEKLDELALKIKNVGYVPNTDFVLHDVEDEQKEQYLFQHSEKL 189
Query: 775 AMAYGLINTGKGIPIRVVKNLRMCSDCHSFAKLVSKLYGREITVRDNNRYHFFKEGFCSC 834
A+A+ LI+T PIRV KNLR+C DCH+ K +S + GREI VRD NR+H K+G CSC
Sbjct: 190 AVAFALISTPNPKPIRVFKNLRVCGDCHTAIKYISMVSGREIVVRDANRFHHMKDGTCSC 369
Query: 835 RDFW 838
D+W
Sbjct: 370 NDYW 381
>AW774626 similar to PIR|T47909|T47 hypothetical protein T20K12.70 -
Arabidopsis thaliana, partial (12%)
Length = 703
Score = 130 bits (327), Expect = 2e-30
Identities = 76/235 (32%), Positives = 121/235 (51%), Gaps = 2/235 (0%)
Frame = +3
Query: 173 LIHFYAACGKVDLGRKVFD--EMLERNVVSWTSLINGYSVVNMAKEAVCLFFEMVEVGVE 230
L+ YA C V +F E +N V WT+++ GY+ +AV F M GVE
Sbjct: 3 LVDMYAKCKCVSEAEFLFKGLEFDRKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVE 182
Query: 231 PNPVTMVCAISACAKLKDLELGKKVCNLMTELGVKSNTLVVNALLDMYMKCGDMYAVREI 290
N T ++AC+ + G++V + + G SN V +AL+DMY KCGD+ + +
Sbjct: 183 CNQYTFPTILTACSSVLARCFGEQVHGFIVKSGFGSNVYVQSALVDMYAKCGDLKNAKNM 362
Query: 291 FDEFSDKNLVMYNTIMSNYVQHGLAGEVLVVLDEMLQKGQRPDKVTMLSTIAACAQLGDL 350
+ D ++V +N++M +V+HGL E L + M + + D T S + C +G +
Sbjct: 363 LETMEDDDVVSWNSLMVGFVRHGLEEEALRLFKNMHGRNMKIDDYTFPSVLNCCV-VGSI 539
Query: 351 SVGKSSHAYVFRNGLERLDNISNAIIDMYMKCGKREAACKVFDSMSNKTVVTWNS 405
+ KS H + + G E +SNA++DMY K G + A VF+ M K V++W S
Sbjct: 540 N-PKSVHGLIIKTGFENYKLVSNALVDMYAKTGDMDCAYTVFEKMLEKDVISWTS 701
Score = 110 bits (274), Expect = 3e-24
Identities = 64/205 (31%), Positives = 115/205 (55%), Gaps = 2/205 (0%)
Frame = +3
Query: 102 LIRGYAASGLCKEAI--FIYLHMIIVMGIVPDNFTFPFLLSACSKIMAFSEGVQVHGVVV 159
++ GYA +G +A+ F Y+H G+ + +TFP +L+ACS ++A G QVHG +V
Sbjct: 102 MVTGYAQNGDGYKAVEFFRYMH---AQGVECNQYTFPTILTACSSVLARCFGEQVHGFIV 272
Query: 160 KMGLVKDLFVANSLIHFYAACGKVDLGRKVFDEMLERNVVSWTSLINGYSVVNMAKEAVC 219
K G +++V ++L+ YA CG + + + + M + +VVSW SL+ G+ + +EA+
Sbjct: 273 KSGFGSNVYVQSALVDMYAKCGDLKNAKNMLETMEDDDVVSWNSLMVGFVRHGLEEEALR 452
Query: 220 LFFEMVEVGVEPNPVTMVCAISACAKLKDLELGKKVCNLMTELGVKSNTLVVNALLDMYM 279
LF M ++ + T ++ C + K V L+ + G ++ LV NAL+DMY
Sbjct: 453 LFKNMHGRNMKIDDYTFPSVLNCC--VVGSINPKSVHGLIIKTGFENYKLVSNALVDMYA 626
Query: 280 KCGDMYAVREIFDEFSDKNLVMYNT 304
K GDM +F++ +K+++ + +
Sbjct: 627 KTGDMDCAYTVFEKMLEKDVISWTS 701
Score = 105 bits (262), Expect = 7e-23
Identities = 72/266 (27%), Positives = 126/266 (47%), Gaps = 2/266 (0%)
Frame = +3
Query: 274 LLDMYMKCGDMYAVREIFD--EFSDKNLVMYNTIMSNYVQHGLAGEVLVVLDEMLQKGQR 331
L+DMY KC + +F EF KN V++ +++ Y Q+G + + M +G
Sbjct: 3 LVDMYAKCKCVSEAEFLFKGLEFDRKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVE 182
Query: 332 PDKVTMLSTIAACAQLGDLSVGKSSHAYVFRNGLERLDNISNAIIDMYMKCGKREAACKV 391
++ T + + AC+ + G+ H ++ ++G + +A++DMY KCG + A +
Sbjct: 183 CNQYTFPTILTACSSVLARCFGEQVHGFIVKSGFGSNVYVQSALVDMYAKCGDLKNAKNM 362
Query: 392 FDSMSNKTVVTWNSLIAGLVRDGELELALRIFGEMPESNLVSWNTMIGAMVQASMFEEAI 451
++M + VV+WNSL+ G VR G E ALR+F
Sbjct: 363 LETMEDDDVVSWNSLMVGFVRHGLEEEALRLF---------------------------- 458
Query: 452 DLLREMQNQGIKGDRVTMVGIASACGYLGALDLAKWIYTYIEKNDIHIDMQLGTALVDMF 511
+ M + +K D T + + C +G+++ K ++ I K + ALVDM+
Sbjct: 459 ---KNMHGRNMKIDDYTFPSVLNCC-VVGSIN-PKSVHGLIIKTGFENYKLVSNALVDMY 623
Query: 512 SRCGDPLNAMRVFENMEKRDVSAWTA 537
++ GD A VFE M ++DV +WT+
Sbjct: 624 AKTGDMDCAYTVFEKMLEKDVISWTS 701
Score = 80.1 bits (196), Expect = 3e-15
Identities = 55/222 (24%), Positives = 105/222 (46%), Gaps = 6/222 (2%)
Frame = +3
Query: 425 EMPESNLVSWNTMIGAMVQASMFEEAIDLLREMQNQGIKGDRVTMVGIASACGYLGALDL 484
E N V W M+ Q +A++ R M QG++ ++ T I +AC + A
Sbjct: 66 EFDRKNHVLWTAMVTGYAQNGDGYKAVEFFRYMHAQGVECNQYTFPTILTACSSVLARCF 245
Query: 485 AKWIYTYIEKNDIHIDMQLGTALVDMFSRCGDPLNAMRVFENMEKRDVSAWTAAIRVKAV 544
+ ++ +I K+ ++ + +ALVDM+++CGD NA + E ME DV +W + +
Sbjct: 246 GEQVHGFIVKSGFGSNVYVQSALVDMYAKCGDLKNAKNMLETMEDDDVVSWNSLMVGFVR 425
Query: 545 EGNAKGAIELFDEMLKQDVKADDFVFVALLTAFSHGGYVDQGRQLFWAMEKIHG--VSPQ 602
G + A+ LF M +++K DD+ F ++L G + +HG +
Sbjct: 426 HGLEEEALRLFKNMHGRNMKIDDYTFPSVLNCCVVGSI---------NPKSVHGLIIKTG 578
Query: 603 IVHY----GCMVDLLGRAGLLEEAFDLMKSMPIKPNDVIWGS 640
+Y +VD+ + G ++ A+ + + M ++ + + W S
Sbjct: 579 FENYKLVSNALVDMYAKTGDMDCAYTVFEKM-LEKDVISWTS 701
>TC83482 weakly similar to GP|9758453|dbj|BAB08982.1 selenium-binding
protein-like {Arabidopsis thaliana}, partial (10%)
Length = 895
Score = 130 bits (326), Expect = 3e-30
Identities = 72/180 (40%), Positives = 103/180 (57%)
Frame = +2
Query: 531 DVSAWTAAIRVKAVEGNAKGAIELFDEMLKQDVKADDFVFVALLTAFSHGGYVDQGRQLF 590
D+ +W A I A G A A+E FD M K D+ FV +L+A H G V++G + F
Sbjct: 2 DLVSWNAIIGGYASHGLATRALEEFDRM-KVVGTPDEVTFVNVLSACVHAGLVEEGEKHF 178
Query: 591 WAMEKIHGVSPQIVHYGCMVDLLGRAGLLEEAFDLMKSMPIKPNDVIWGSFLAACRKHKN 650
M +G+ ++ HY CMVDL GRAG +EA +L+K+MP +P+ V+WG+ LAAC H N
Sbjct: 179 TDMLTKYGIQAEMEHYSCMVDLYGRAGRFDEAENLIKNMPFEPDVVLWGALLAACGLHSN 358
Query: 651 VEFANYADEKITQLAPEKVGIHVLLSNIYASAGKWNDVARVRLQMKEKGFQKVAGSSSIE 710
+E YA E+I +L + +LS I G W+ V +R MKE+G +K S +E
Sbjct: 359 LELGEYAAERIRRLESSHPVSYSVLSKIQGEKGVWSSVNELRDTMKERGIKKQTAISWVE 538
Score = 59.3 bits (142), Expect = 6e-09
Identities = 44/171 (25%), Positives = 80/171 (46%), Gaps = 5/171 (2%)
Frame = +2
Query: 298 NLVMYNTIMSNYVQHGLAGEVLVVLDEMLQKGQRPDKVTMLSTIAACAQLGDLSVGKSSH 357
+LV +N I+ Y HGLA L D M G PD+VT ++ ++AC G + G+
Sbjct: 2 DLVSWNAIIGGYASHGLATRALEEFDRMKVVGT-PDEVTFVNVLSACVHAGLVEEGEKHF 178
Query: 358 A-YVFRNGLERLDNISNAIIDMYMKCGKREAACKVFDSMS-NKTVVTWNSLIAGLVRDGE 415
+ + G++ + ++D+Y + G+ + A + +M VV W +L+A
Sbjct: 179 TDMLTKYGIQAEMEHYSCMVDLYGRAGRFDEAENLIKNMPFEPDVVLWGALLAACGLHSN 358
Query: 416 LEL---ALRIFGEMPESNLVSWNTMIGAMVQASMFEEAIDLLREMQNQGIK 463
LEL A + S+ VS++ + + ++ +L M+ +GIK
Sbjct: 359 LELGEYAAERIRRLESSHPVSYSVLSKIQGEKGVWSSVNELRDTMKERGIK 511
Score = 46.2 bits (108), Expect = 5e-05
Identities = 27/87 (31%), Positives = 50/87 (57%), Gaps = 1/87 (1%)
Frame = +2
Query: 197 NVVSWTSLINGYSVVNMAKEAVCLFFEMVEVGVEPNPVTMVCAISACAKLKDLELGKK-V 255
++VSW ++I GY+ +A A+ F M VG P+ VT V +SAC +E G+K
Sbjct: 2 DLVSWNAIIGGYASHGLATRALEEFDRMKVVGT-PDEVTFVNVLSACVHAGLVEEGEKHF 178
Query: 256 CNLMTELGVKSNTLVVNALLDMYMKCG 282
+++T+ G+++ + ++D+Y + G
Sbjct: 179 TDMLTKYGIQAEMEHYSCMVDLYGRAG 259
Score = 43.5 bits (101), Expect = 3e-04
Identities = 32/112 (28%), Positives = 53/112 (46%), Gaps = 2/112 (1%)
Frame = +2
Query: 96 LYTCNTLIRGYAASGLCKEAIFIYLHMIIVMGIVPDNFTFPFLLSACSKIMAFSEGVQ-V 154
L + N +I GYA+ GL A+ + M +V PD TF +LSAC EG +
Sbjct: 5 LVSWNAIIGGYASHGLATRALEEFDRMKVVG--TPDEVTFVNVLSACVHAGLVEEGEKHF 178
Query: 155 HGVVVKMGLVKDLFVANSLIHFYAACGKVDLGRKVFDEM-LERNVVSWTSLI 205
++ K G+ ++ + ++ Y G+ D + M E +VV W +L+
Sbjct: 179 TDMLTKYGIQAEMEHYSCMVDLYGRAGRFDEAENLIKNMPFEPDVVLWGALL 334
>BE202782 weakly similar to GP|22093801|dbj hypothetical protein~predicted by
GlimmerM~similar to Arabidopsis thaliana chromosome3
At3g23330, partial (13%)
Length = 548
Score = 127 bits (319), Expect = 2e-29
Identities = 57/93 (61%), Positives = 72/93 (77%)
Frame = +2
Query: 746 GYVPDTTNVLVDVDEQEKEHLLSRHSEKLAMAYGLINTGKGIPIRVVKNLRMCSDCHSFA 805
G +P T +VL DV+EQ+KE +L HSEKLA+ GLINT G P++V+KNLR+C DCH+
Sbjct: 2 GCLPMTKSVLQDVEEQDKEQILCGHSEKLAVVLGLINTSPGQPLQVIKNLRICDDCHAVI 181
Query: 806 KLVSKLYGREITVRDNNRYHFFKEGFCSCRDFW 838
K++S+L GREI VRD NR+H FKEG CSC DFW
Sbjct: 182 KVISRLEGREIFVRDTNRFHHFKEGVCSCADFW 280
>BI308000 similar to GP|7363286|dbj| Similar to Arabidopsis thaliana DNA
chromosome 4 BAC clone F4I10; vegetative storage
protein., partial (12%)
Length = 771
Score = 124 bits (310), Expect = 2e-28
Identities = 58/109 (53%), Positives = 75/109 (68%)
Frame = +3
Query: 730 QIGLMLQEINCRISQVGYVPDTTNVLVDVDEQEKEHLLSRHSEKLAMAYGLINTGKGIPI 789
+I L+ + +I GY+PDT N + DV+E+ KE LLS HSE+LA+A+GL+NT G PI
Sbjct: 12 EIYAFLEALGDKIRDAGYIPDT-NSIHDVEEKVKEQLLSSHSERLAIAFGLLNTNHGTPI 188
Query: 790 RVVKNLRMCSDCHSFAKLVSKLYGREITVRDNNRYHFFKEGFCSCRDFW 838
V KNLR+C DCH K +S + GREI VRD R+H FK G CSC D+W
Sbjct: 189 HVRKNLRVCGDCHDVTKYISLVTGREIIVRDLRRFHHFKNGICSCGDYW 335
>AW696172 weakly similar to PIR|H96620|H96 hypothetical protein F23H11.3
[imported] - Arabidopsis thaliana, partial (19%)
Length = 649
Score = 115 bits (288), Expect = 7e-26
Identities = 66/151 (43%), Positives = 93/151 (60%), Gaps = 4/151 (2%)
Frame = +2
Query: 97 YTCNTLIRGYAASGLCKE-AIFIYLHMIIVMG--IVPDNFTFPFLLSACSKIMAFSEGVQ 153
+T N LI+ Y+ S L K+ AI +Y +I + PD T+PF+L AC+ + + EG Q
Sbjct: 164 FTWNILIQSYSKSTLHKQKAILLYKAIITEQENELFPDKHTYPFVLKACAYLFSLFEGKQ 343
Query: 154 VHGVVVKMGLVKDLFVANSLIHFYAACGKVDLGRKVFDEMLE-RNVVSWTSLINGYSVVN 212
VH V+K+G D ++ NSLIHFYA+CG ++ RKVFD M E RNVVSW +I+ Y+ V
Sbjct: 344 VHAHVLKLGFELDTYICNSLIHFYASCGYLETARKVFDRMCEWRNVVSWNVMIDSYAKVG 523
Query: 213 MAKEAVCLFFEMVEVGVEPNPVTMVCAISAC 243
+ +F EM++V EP+ TM I AC
Sbjct: 524 DYDIVLIMFCEMMKV-YEPDCYTMQSVIRAC 613
Score = 74.3 bits (181), Expect = 2e-13
Identities = 48/153 (31%), Positives = 78/153 (50%), Gaps = 5/153 (3%)
Frame = +2
Query: 197 NVVSWTSLINGYSVVNMAKEAVCLFFEMVEVGVE----PNPVTMVCAISACAKLKDLELG 252
N +W LI YS + K+ L ++ + E P+ T + ACA L L G
Sbjct: 158 NSFTWNILIQSYSKSTLHKQKAILLYKAIITEQENELFPDKHTYPFVLKACAYLFSLFEG 337
Query: 253 KKVCNLMTELGVKSNTLVVNALLDMYMKCGDMYAVREIFDEFSD-KNLVMYNTIMSNYVQ 311
K+V + +LG + +T + N+L+ Y CG + R++FD + +N+V +N ++ +Y +
Sbjct: 338 KQVHAHVLKLGFELDTYICNSLIHFYASCGYLETARKVFDRMCEWRNVVSWNVMIDSYAK 517
Query: 312 HGLAGEVLVVLDEMLQKGQRPDKVTMLSTIAAC 344
G VL++ EM+ K PD TM S I AC
Sbjct: 518 VGDYDIVLIMFCEMM-KVYEPDCYTMQSVIRAC 613
Score = 70.1 bits (170), Expect = 3e-12
Identities = 45/134 (33%), Positives = 68/134 (50%), Gaps = 5/134 (3%)
Frame = +2
Query: 298 NLVMYNTIMSNYVQHGLAGEVLVVLDEMLQKGQR----PDKVTMLSTIAACAQLGDLSVG 353
N +N ++ +Y + L + ++L + + Q PDK T + ACA L L G
Sbjct: 158 NSFTWNILIQSYSKSTLHKQKAILLYKAIITEQENELFPDKHTYPFVLKACAYLFSLFEG 337
Query: 354 KSSHAYVFRNGLERLDNISNAIIDMYMKCGKREAACKVFDSMSN-KTVVTWNSLIAGLVR 412
K HA+V + G E I N++I Y CG E A KVFD M + VV+WN +I +
Sbjct: 338 KQVHAHVLKLGFELDTYICNSLIHFYASCGYLETARKVFDRMCEWRNVVSWNVMIDSYAK 517
Query: 413 DGELELALRIFGEM 426
G+ ++ L +F EM
Sbjct: 518 VGDYDIVLIMFCEM 559
Score = 52.0 bits (123), Expect = 9e-07
Identities = 35/136 (25%), Positives = 67/136 (48%), Gaps = 5/136 (3%)
Frame = +2
Query: 430 NLVSWNTMIGAMVQASMFEEAIDLLREM----QNQGIKGDRVTMVGIASACGYLGALDLA 485
N +WN +I + ++++ ++ LL + Q + D+ T + AC YL +L
Sbjct: 158 NSFTWNILIQSYSKSTLHKQKAILLYKAIITEQENELFPDKHTYPFVLKACAYLFSLFEG 337
Query: 486 KWIYTYIEKNDIHIDMQLGTALVDMFSRCGDPLNAMRVFENM-EKRDVSAWTAAIRVKAV 544
K ++ ++ K +D + +L+ ++ CG A +VF+ M E R+V +W I A
Sbjct: 338 KQVHAHVLKLGFELDTYICNSLIHFYASCGYLETARKVFDRMCEWRNVVSWNVMIDSYAK 517
Query: 545 EGNAKGAIELFDEMLK 560
G+ + +F EM+K
Sbjct: 518 VGDYDIVLIMFCEMMK 565
>TC84835 weakly similar to PIR|T02328|T02328 hypothetical protein At2g34400
[imported] - Arabidopsis thaliana, partial (5%)
Length = 647
Score = 113 bits (283), Expect = 3e-25
Identities = 63/199 (31%), Positives = 114/199 (56%), Gaps = 1/199 (0%)
Frame = +1
Query: 64 NKLIAACVQMGTHESLNYALNAFKEDEGTKCSLYTCNTLIRGYAASGLCKEAIFIYLHMI 123
+KL + + + + +Y+L+ F SL+ N LI+ + + I ++ + +
Sbjct: 10 HKLFSVSIHLNNNNYFHYSLSIFNHT--LHPSLFLYNLLIKSFFKRNSFQTLISLF-NQL 180
Query: 124 IVMGIVPDNFTFPFLLSACSKIMAFSEGVQVHGVVVKMGLVKDLFVANSLIHFYAACGKV 183
+ G+ PDN+T+PF+L A + I F +G ++H V K GL D +V+NS + YA G++
Sbjct: 181 RLNGLYPDNYTYPFVLKAVAFIADFRQGTKIHAFVFKTGLDSDYYVSNSFMDMYAELGRI 360
Query: 184 DLGRKVFDEMLERNVVSWTSLINGYSVVNMAKEAVCLFFEM-VEVGVEPNPVTMVCAISA 242
D RK+FDE+ ER+ VSW +I+G +EAV +F M V+ + + T+V +++A
Sbjct: 361 DFVRKLFDEISERDSVSWNVMISGCVKCRRFEEAVEVFQRMRVDSNEKISEATVVSSLTA 540
Query: 243 CAKLKDLELGKKVCNLMTE 261
CA +++E+GK++ + E
Sbjct: 541 CAASRNVEVGKEIHGFIIE 597
Score = 78.6 bits (192), Expect = 9e-15
Identities = 46/179 (25%), Positives = 92/179 (50%), Gaps = 1/179 (0%)
Frame = +1
Query: 189 VFDEMLERNVVSWTSLINGYSVVNMAKEAVCLFFEMVEVGVEPNPVTMVCAISACAKLKD 248
+F+ L ++ + LI + N + + LF ++ G+ P+ T + A A + D
Sbjct: 73 IFNHTLHPSLFLYNLLIKSFFKRNSFQTLISLFNQLRLNGLYPDNYTYPFVLKAVAFIAD 252
Query: 249 LELGKKVCNLMTELGVKSNTLVVNALLDMYMKCGDMYAVREIFDEFSDKNLVMYNTIMSN 308
G K+ + + G+ S+ V N+ +DMY + G + VR++FDE S+++ V +N ++S
Sbjct: 253 FRQGTKIHAFVFKTGLDSDYYVSNSFMDMYAELGRIDFVRKLFDEISERDSVSWNVMISG 432
Query: 309 YVQHGLAGEVLVVLDEM-LQKGQRPDKVTMLSTIAACAQLGDLSVGKSSHAYVFRNGLE 366
V+ E + V M + ++ + T++S++ ACA ++ VGK H ++ L+
Sbjct: 433 CVKCRRFEEAVEVFQRMRVDSNEKISEATVVSSLTACAASRNVEVGKEIHGFIIEKELD 609
Score = 78.2 bits (191), Expect = 1e-14
Identities = 47/180 (26%), Positives = 88/180 (48%), Gaps = 16/180 (8%)
Frame = +1
Query: 290 IFDEFSDKNLVMYNTIMSNYVQHGLAGEVLVVLDEMLQKGQRPDKVTMLSTIAACAQLGD 349
IF+ +L +YN ++ ++ + ++ + +++ G PD T + A A + D
Sbjct: 73 IFNHTLHPSLFLYNLLIKSFFKRNSFQTLISLFNQLRLNGLYPDNYTYPFVLKAVAFIAD 252
Query: 350 LSVGKSSHAYVFRNGLERLDNISNAIIDMYMKCGKREAACKVFDSMSNKTVVTWNSLIAG 409
G HA+VF+ GL+ +SN+ +DMY + G+ + K+FD +S + V+WN +I+G
Sbjct: 253 FRQGTKIHAFVFKTGLDSDYYVSNSFMDMYAELGRIDFVRKLFDEISERDSVSWNVMISG 432
Query: 410 LVRDGELELALRIFGEM--------PESNLV--------SWNTMIGAMVQASMFEEAIDL 453
V+ E A+ +F M E+ +V S N +G + + E+ +DL
Sbjct: 433 CVKCRRFEEAVEVFQRMRVDSNEKISEATVVSSLTACAASRNVEVGKEIHGFIIEKELDL 612
>TC83657 weakly similar to GP|11994382|dbj|BAB02341.
gb|AAB82628.1~gene_id:MIL23.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1299
Score = 110 bits (276), Expect = 2e-24
Identities = 58/140 (41%), Positives = 89/140 (63%), Gaps = 1/140 (0%)
Frame = +3
Query: 571 VALLTAFSHGGYVDQGRQLFWAMEKIHGVSPQIVHYGCMVDLLGRAGLLEEAFDLMKSMP 630
+ +L+A +HGG + + ++ ME+ +G+ I HYGCMVDLLGRAG L+EA++L+K MP
Sbjct: 78 LXVLSACAHGGLMSEALEVISKMEE-YGIEMGIRHYGCMVDLLGRAGKLKEAYELIKRMP 254
Query: 631 IKPNDVIWGSFLAACRKHKNVEFANYADEKITQLAPEKVGIH-VLLSNIYASAGKWNDVA 689
+KPN+ + G+ + AC H +++ A + I + V H VLLSNIYA++ KW
Sbjct: 255 MKPNETVLGAMIGACWIHSDMKMAEQVMKMIGADSAACVNSHNVLLSNIYAASEKWEKAE 434
Query: 690 RVRLQMKEKGFQKVAGSSSI 709
+R M + G +K+ G SSI
Sbjct: 435 MIRSSMVDGGSEKIPGYSSI 494
>AW257410 weakly similar to GP|10177533|db selenium-binding protein-like
{Arabidopsis thaliana}, partial (21%)
Length = 724
Score = 109 bits (272), Expect = 5e-24
Identities = 64/226 (28%), Positives = 118/226 (51%), Gaps = 2/226 (0%)
Frame = +1
Query: 243 CAKLKDLELGKKVCNLMTELGVKSNTLVVNALLDMY--MKCGDMYAVREIFDEFSDKNLV 300
C+ + +L K++ + G + L V+ LL Y M+ ++ R +FD S N V
Sbjct: 7 CSNIGEL---KQIYVQFLKKGTIRHKLTVSRLLTTYASMEFSNLTYARMVFDRISSPNTV 177
Query: 301 MYNTIMSNYVQHGLAGEVLVVLDEMLQKGQRPDKVTMLSTIAACAQLGDLSVGKSSHAYV 360
M+NT++ Y E L++ +ML + T + AC+ L L+ H +
Sbjct: 178 MWNTMIRAYSNSNDPEEALLLYHQMLHHSIPHNAYTFPFLLKACSALSALAETHQIHVQI 357
Query: 361 FRNGLERLDNISNAIIDMYMKCGKREAACKVFDSMSNKTVVTWNSLIAGLVRDGELELAL 420
+ G +N+++ +Y G ++A +FD + ++ +V+WN++I G ++ G +E+A
Sbjct: 358 IKRGFGSEVYATNSLLRVYAISGSIKSAHVLFDLLPSRDIVSWNTMIDGYIKCGNVEMAY 537
Query: 421 RIFGEMPESNLVSWNTMIGAMVQASMFEEAIDLLREMQNQGIKGDR 466
+IF MPE N++SW +MI V+ M ++A+ LL++M IK D+
Sbjct: 538 KIFQAMPEKNVISWTSMIVGFVRTGMHKKALSLLQQMLVARIKPDK 675
Score = 105 bits (263), Expect = 6e-23
Identities = 67/228 (29%), Positives = 110/228 (47%), Gaps = 35/228 (15%)
Frame = +1
Query: 40 QTCKTLIELKQLHCNMLKKGVFN----INKLIAACVQMGTHESLNYALNAFKEDEGTKCS 95
+ C + ELKQ++ LKKG +++L+ M +L YA F
Sbjct: 1 ERCSNIGELKQIYVQFLKKGTIRHKLTVSRLLTTYASM-EFSNLTYARMVFDRISSPNTV 177
Query: 96 LYTCNTLIRGYAASGLCKEAIFIYLHMIIVMGIVPDNFTFPFLLSACSKIMAFSEGVQVH 155
++ NT+IR Y+ S +EA+ +Y H ++ I + +TFPFLL ACS + A +E Q+H
Sbjct: 178 MW--NTMIRAYSNSNDPEEALLLY-HQMLHHSIPHNAYTFPFLLKACSALSALAETHQIH 348
Query: 156 GVVVKMGLVKDLFVANSLIHFYA-------------------------------ACGKVD 184
++K G +++ NSL+ YA CG V+
Sbjct: 349 VQIIKRGFGSEVYATNSLLRVYAISGSIKSAHVLFDLLPSRDIVSWNTMIDGYIKCGNVE 528
Query: 185 LGRKVFDEMLERNVVSWTSLINGYSVVNMAKEAVCLFFEMVEVGVEPN 232
+ K+F M E+NV+SWTS+I G+ M K+A+ L +M+ ++P+
Sbjct: 529 MAYKIFQAMPEKNVISWTSMIVGFVRTGMHKKALSLLQQMLVARIKPD 672
Score = 73.6 bits (179), Expect = 3e-13
Identities = 54/226 (23%), Positives = 97/226 (42%), Gaps = 33/226 (14%)
Frame = +1
Query: 142 CSKIMAFSEGVQVHGVVVKMGLVKDLFVANSLIHFYAAC--GKVDLGRKVFDEMLERNVV 199
CS I E Q++ +K G ++ + L+ YA+ + R VFD + N V
Sbjct: 7 CSNI---GELKQIYVQFLKKGTIRHKLTVSRLLTTYASMEFSNLTYARMVFDRISSPNTV 177
Query: 200 SWTSLINGYSVVNMAKEAVCLFFEMVEVGVEPNPVTMVCAISACAKLKDLELGKKVCNLM 259
W ++I YS N +EA+ L+ +M+ + N T + AC+ L L ++ +
Sbjct: 178 MWNTMIRAYSNSNDPEEALLLYHQMLHHSIPHNAYTFPFLLKACSALSALAETHQIHVQI 357
Query: 260 TELGVKSNTLVVNALL-------------------------------DMYMKCGDMYAVR 288
+ G S N+LL D Y+KCG++
Sbjct: 358 IKRGFGSEVYATNSLLRVYAISGSIKSAHVLFDLLPSRDIVSWNTMIDGYIKCGNVEMAY 537
Query: 289 EIFDEFSDKNLVMYNTIMSNYVQHGLAGEVLVVLDEMLQKGQRPDK 334
+IF +KN++ + +++ +V+ G+ + L +L +ML +PDK
Sbjct: 538 KIFQAMPEKNVISWTSMIVGFVRTGMHKKALSLLQQMLVARIKPDK 675
Score = 58.9 bits (141), Expect = 8e-09
Identities = 40/148 (27%), Positives = 69/148 (46%)
Frame = +1
Query: 416 LELALRIFGEMPESNLVSWNTMIGAMVQASMFEEAIDLLREMQNQGIKGDRVTMVGIASA 475
L A +F + N V WNTMI A ++ EEA+ L +M + I + T + A
Sbjct: 127 LTYARMVFDRISSPNTVMWNTMIRAYSNSNDPEEALLLYHQMLHHSIPHNAYTFPFLLKA 306
Query: 476 CGYLGALDLAKWIYTYIEKNDIHIDMQLGTALVDMFSRCGDPLNAMRVFENMEKRDVSAW 535
C L AL I+ I K ++ +L+ +++ G +A +F+ + RD+ +W
Sbjct: 307 CSALSALAETHQIHVQIIKRGFGSEVYATNSLLRVYAISGSIKSAHVLFDLLPSRDIVSW 486
Query: 536 TAAIRVKAVEGNAKGAIELFDEMLKQDV 563
I GN + A ++F M +++V
Sbjct: 487 NTMIDGYIKCGNVEMAYKIFQAMPEKNV 570
>BG645370 weakly similar to PIR|T00405|T004 hypothetical protein At2g44880
[imported] - Arabidopsis thaliana, partial (11%)
Length = 784
Score = 108 bits (270), Expect = 8e-24
Identities = 53/138 (38%), Positives = 85/138 (61%)
Frame = -2
Query: 570 FVALLTAFSHGGYVDQGRQLFWAMEKIHGVSPQIVHYGCMVDLLGRAGLLEEAFDLMKSM 629
F A+LTA +H G++D+G + F M +G+SP I Y C++DL R G L +A DLM+ M
Sbjct: 783 FTAVLTACNHAGFIDKGEEYFNKMITNYGLSPDIDIYACLIDLYARNGNLRKARDLMEEM 604
Query: 630 PIKPNDVIWGSFLAACRKHKNVEFANYADEKITQLAPEKVGIHVLLSNIYASAGKWNDVA 689
P PN +IW SFL+AC+ + +VE A ++ ++ P ++ L++IY + G WN+ +
Sbjct: 603 PYDPNCIIWSSFLSACKIYGDVELGREAAIQLIKMEPCNAAPYLTLAHIYTTKGLWNEAS 424
Query: 690 RVRLQMKEKGFQKVAGSS 707
VR M+++ +K G S
Sbjct: 423 EVRSLMQQRVKRKPPGWS 370
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,590,360
Number of Sequences: 36976
Number of extensions: 329999
Number of successful extensions: 2144
Number of sequences better than 10.0: 141
Number of HSP's better than 10.0 without gapping: 1811
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2075
length of query: 838
length of database: 9,014,727
effective HSP length: 104
effective length of query: 734
effective length of database: 5,169,223
effective search space: 3794209682
effective search space used: 3794209682
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146852.15


