

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146842.6 - phase: 0
(535 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
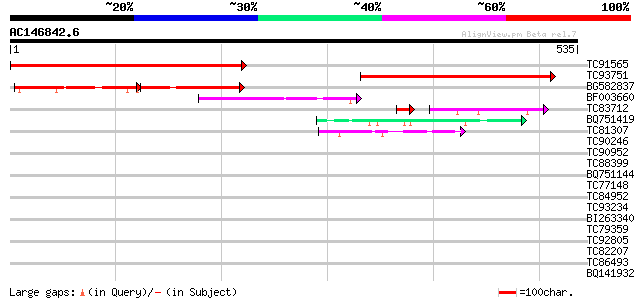
Score E
Sequences producing significant alignments: (bits) Value
TC91565 similar to GP|9293900|dbj|BAB01803.1 emb|CAB62463.1~gene... 434 e-122
TC93751 similar to GP|9293900|dbj|BAB01803.1 emb|CAB62463.1~gene... 375 e-104
BG582837 similar to GP|9293900|dbj emb|CAB62463.1~gene_id:MVE11.... 123 8e-59
BF003660 homologue to PIR|T46236|T462 hypothetical protein T9C5.... 106 3e-23
TC83712 similar to PIR|T46236|T46236 hypothetical protein T9C5.1... 70 7e-15
BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania maj... 44 1e-04
TC81307 weakly similar to GP|4335772|gb|AAD17449.1| unknown prot... 42 6e-04
TC90246 similar to PIR|T00872|T00872 probable protein kinase At2... 41 0.001
TC90952 similar to GP|15724316|gb|AAL06551.1 AT4g34150/F28A23_90... 41 0.001
TC88399 weakly similar to GP|18491217|gb|AAL69511.1 putative AMP... 39 0.004
BQ751144 weakly similar to PIR|PQ0479|PQ04 pistil extensin-like ... 39 0.005
TC77148 ENOD20 39 0.005
TC84952 similar to PIR|T51759|T51759 [glutamate--ammonia-ligase]... 38 0.011
TC93234 weakly similar to GP|21751020|dbj|BAC03887. unnamed prot... 37 0.015
BI263340 weakly similar to PIR|T49223|T492 uclacyanin 3 [importe... 37 0.025
TC79359 weakly similar to PIR|S49915|S49915 extensin-like protei... 36 0.042
TC92805 similar to PIR|A86459|A86459 unknown protein 82634-8124... 36 0.042
TC82207 similar to GP|14517356|gb|AAK62569.1 AT4g15540/dl3810w {... 36 0.042
TC86493 similar to GP|18654317|gb|AAL77589.1 ribose-5-phosphate ... 35 0.055
BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 35 0.055
>TC91565 similar to GP|9293900|dbj|BAB01803.1
emb|CAB62463.1~gene_id:MVE11.13~similar to unknown
protein {Arabidopsis thaliana}, partial (14%)
Length = 701
Score = 434 bits (1116), Expect = e-122
Identities = 223/223 (100%), Positives = 223/223 (100%)
Frame = +2
Query: 1 MTTTSSHAQTEAAKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSV 60
MTTTSSHAQTEAAKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSV
Sbjct: 32 MTTTSSHAQTEAAKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSV 211
Query: 61 RPRDKWFNLALRECPTALENTDLWRHSNLQPIVVDVVLVHRNLSFSPKVRSFVKERNPFE 120
RPRDKWFNLALRECPTALENTDLWRHSNLQPIVVDVVLVHRNLSFSPKVRSFVKERNPFE
Sbjct: 212 RPRDKWFNLALRECPTALENTDLWRHSNLQPIVVDVVLVHRNLSFSPKVRSFVKERNPFE 391
Query: 121 EFGGGSEQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLY 180
EFGGGSEQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLY
Sbjct: 392 EFGGGSEQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLY 571
Query: 181 ATVRLLPAFKIFKELSSSANVSAFSLAHRVSTFFEPFTTKQEA 223
ATVRLLPAFKIFKELSSSANVSAFSLAHRVSTFFEPFTTKQEA
Sbjct: 572 ATVRLLPAFKIFKELSSSANVSAFSLAHRVSTFFEPFTTKQEA 700
>TC93751 similar to GP|9293900|dbj|BAB01803.1
emb|CAB62463.1~gene_id:MVE11.13~similar to unknown
protein {Arabidopsis thaliana}, partial (7%)
Length = 1166
Score = 375 bits (962), Expect = e-104
Identities = 184/185 (99%), Positives = 184/185 (99%), Gaps = 1/185 (0%)
Frame = +1
Query: 332 HT-LVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSL 390
HT LVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSL
Sbjct: 1 HTHLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSL 180
Query: 391 GSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSC 450
GSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSC
Sbjct: 181 GSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSC 360
Query: 451 TTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSRSYQDEFDDTDFTCPFDVDDDDTT 510
TTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSRSYQDEFDDTDFTCPFDVDDDDTT
Sbjct: 361 TTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSRSYQDEFDDTDFTCPFDVDDDDTT 540
Query: 511 DPGSR 515
DPGSR
Sbjct: 541 DPGSR 555
>BG582837 similar to GP|9293900|dbj emb|CAB62463.1~gene_id:MVE11.13~similar
to unknown protein {Arabidopsis thaliana}, partial (22%)
Length = 836
Score = 123 bits (308), Expect(2) = 8e-59
Identities = 61/98 (62%), Positives = 79/98 (80%)
Frame = +1
Query: 124 GGSEQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYATV 183
G ++E+I+ERW +QY+++KIKD S TRRS+N FLQNLYKKSTLLLRSLYATV
Sbjct: 532 GVEAKSERIIERWFVQYKNRKIKDSDSG-----TRRSNNYFLQNLYKKSTLLLRSLYATV 696
Query: 184 RLLPAFKIFKELSSSANVSAFSLAHRVSTFFEPFTTKQ 221
RLLPA++IF++L+SSA + F+LA RVS+F EPFT K+
Sbjct: 697 RLLPAYRIFRDLNSSAKIHPFTLARRVSSFVEPFTRKE 810
Score = 122 bits (307), Expect(2) = 8e-59
Identities = 80/139 (57%), Positives = 93/139 (66%), Gaps = 19/139 (13%)
Frame = +3
Query: 5 SSH----AQTEAAKTEQIITEFFAKSLHIIIESRALSASSRN----FSTLSSPSSTSSSS 56
SSH AQ++ AK EQIITEFFAKSLHII+ESR L ASSRN + + SSP S SSSS
Sbjct: 138 SSHHHGSAQSDNAKIEQIITEFFAKSLHIILESRTLYASSRNSYGGYKSDSSPCS-SSSS 314
Query: 57 SSSVRPRDKWFNLALRECPTALENTDLWRHSNLQPIVVDVVLVHRNLSFSPKV------- 109
SSSVR RDKWFNLALRECP ALEN +NL I++DVVLV+++L + P
Sbjct: 315 SSSVRSRDKWFNLALRECPAALENI-----NNLDSIIIDVVLVNKSLGWDPMTPKRVILR 479
Query: 110 RSFVKERNPF----EEFGG 124
S +KER P EE GG
Sbjct: 480 SSSLKERYPLCCHGEELGG 536
>BF003660 homologue to PIR|T46236|T462 hypothetical protein T9C5.180 -
Arabidopsis thaliana, partial (3%)
Length = 556
Score = 106 bits (264), Expect = 3e-23
Identities = 64/157 (40%), Positives = 93/157 (58%), Gaps = 3/157 (1%)
Frame = +3
Query: 179 LYATVRLLPAFKIFKELSSSANVSAFSLAHRVSTFFEPFTTKQEAEMLKFVFTPVDTNSG 238
LY+ +RLLP IF++LS++ + F + ++VS+F +PF+ ++ M +++FTP+D G
Sbjct: 3 LYSQMRLLPIHIIFRQLSATNHSCNFDIVYKVSSFSDPFSREEGGMMGEYIFTPIDALPG 182
Query: 239 KLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFP-SLPVVRMPSHGSSPS 297
+L LSV YR +SD + + T L ++I DYVGSP TD L FP S+ VR P PS
Sbjct: 183 RLSLSVTYRTTLSDFNLQCLT-LPTKIIADYVGSPNTDPLRYFPSSVKGVRAP-----PS 344
Query: 298 SLPFSRRHSWSYENCRASPPAVN--YYSPSPTHSESH 332
S P R HSW+ +A+P N Y P H SH
Sbjct: 345 SSPLERPHSWTPGFHKAAPFVQNHQYVGSPPAHRGSH 455
>TC83712 similar to PIR|T46236|T46236 hypothetical protein T9C5.180 -
Arabidopsis thaliana, partial (13%)
Length = 672
Score = 70.5 bits (171), Expect(2) = 7e-15
Identities = 46/124 (37%), Positives = 68/124 (54%), Gaps = 12/124 (9%)
Frame = +1
Query: 397 TLLRSESAPVNIPNAEVTYSPGHTS-------RQNLPPSTPIRISRCTSETE---RSRNL 446
T +RSE+APV IP+ + S + S R +LPP +P R + E+ RS
Sbjct: 85 TRMRSETAPVTIPHPMMGKSSRNLSPNFLDPNRNSLPPLSPRRNDTSSHESPSGIRSFRK 264
Query: 447 MQSCTTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSR--SYQDEFDDTDFTCPFDV 504
++S +K+ K+ G ++ + SP+I SR+SS S+QD+ DD DF+CPFDV
Sbjct: 265 IESLKIGQKIVRDSKDDSGRFSGLLSSSDSPRIGASRTSSSRLSFQDDLDDGDFSCPFDV 444
Query: 505 DDDD 508
DD D
Sbjct: 445 DDVD 456
Score = 28.1 bits (61), Expect(2) = 7e-15
Identities = 12/17 (70%), Positives = 13/17 (75%)
Frame = +3
Query: 366 FDEYYPSPNFSPSPSPS 382
+DEY SP FS SPSPS
Sbjct: 3 YDEYQLSPPFSSSPSPS 53
>BQ751419 weakly similar to GP|21629340|gb L509.2 {Leishmania major}, partial
(1%)
Length = 766
Score = 44.3 bits (103), Expect = 1e-04
Identities = 61/223 (27%), Positives = 86/223 (38%), Gaps = 25/223 (11%)
Frame = +1
Query: 290 PSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNA------SSQGY 343
P SSPS+ P S ++PP +PSPT + TL N ++Q
Sbjct: 46 PPASSSPSARP-----SPPPTTASSTPPGAP--TPSPTPTRRATLNGNTPAPASPTAQAT 204
Query: 344 CP--PSSLPPHPSEMSLLHKKNVNFDEYYP----------SPNFS---PSPSPSSSSPIY 388
P PS+ P PS + + + P SP+ S P+P+PS+S
Sbjct: 205 PPTGPSTAAPSPSTCTTTGPTSSSTSASSPAATSPRTTSPSPHSSGTPPAPAPSASRSST 384
Query: 389 SLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPS----TPIRISRCTSETERSR 444
S A S S+P P T +P SRQ PPS + SRCT R+
Sbjct: 385 SRPPRARARRAASRSSPWATPARACTTAPTSASRQTPPPSPRTAAKPKASRCTPSRSRAP 564
Query: 445 NLMQSCTTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSR 487
T P KM + + + A ++P + R SSR
Sbjct: 565 ------TEPSKMAAAAAAAGTRATRPRATAAAPHRAALRLSSR 675
>TC81307 weakly similar to GP|4335772|gb|AAD17449.1| unknown protein
{Arabidopsis thaliana}, partial (31%)
Length = 1171
Score = 42.0 bits (97), Expect = 6e-04
Identities = 48/148 (32%), Positives = 68/148 (45%), Gaps = 9/148 (6%)
Frame = +2
Query: 292 HGSSPSSLPFSRRHSWSY---ENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSS 348
H P L + + +SY +C SP A+ SP+ + + S + SN S Q P SS
Sbjct: 323 HLKQPLLLIYLSPYHFSYCLSSSCSFSPSAL*PPSPTASSTASSAVPSNFSLQS--PLSS 496
Query: 349 LP-----PHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSES 403
P P PS + S + SPSPSPSS+SP SL S A L S
Sbjct: 497 PPFFLSLPQPSSPT-------------SSSSPSPSPSPSSASPSPSLKSFA----LSPPS 625
Query: 404 APVNIP-NAEVTYSPGHTSRQNLPPSTP 430
+ ++ P ++ + S GH+ PPS+P
Sbjct: 626 SFLHSPFSSSTSVSHGHS-----PPSSP 694
Score = 31.2 bits (69), Expect = 1.0
Identities = 25/70 (35%), Positives = 33/70 (46%), Gaps = 2/70 (2%)
Frame = +2
Query: 281 FPSLPVVRMPSHGSSPSSLPF--SRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNA 338
F SLP P+ SSPS P S S S ++ SPP+ +SP + S +
Sbjct: 506 FLSLPQPSSPTSSSSPSPSPSPSSASPSPSLKSFALSPPSSFLHSPFSS--------STS 661
Query: 339 SSQGYCPPSS 348
S G+ PPSS
Sbjct: 662 VSHGHSPPSS 691
>TC90246 similar to PIR|T00872|T00872 probable protein kinase At2g45590
[imported] - Arabidopsis thaliana, partial (28%)
Length = 1451
Score = 41.2 bits (95), Expect = 0.001
Identities = 39/111 (35%), Positives = 53/111 (47%), Gaps = 15/111 (13%)
Frame = -1
Query: 290 PSHGSSPSSLP---FSRRHSWSYENC-RASPPAV----NYYSPSPTHSES-HTLVSNASS 340
P H + PS++P FS S S++ R S P + + SP +HS+S H L+ SS
Sbjct: 410 PLHNAPPSTIPSPTFSSLSSSSHQQFPRPSIPTIPISASSSSPFSSHSQSYHPLIH--SS 237
Query: 341 QGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPS------PNFSPSPSPSSSS 385
P S P H S LH+ N +YP PN+S + SPS SS
Sbjct: 236 HFSLPFQSNPSHNPSPSYLHQLASNPQPHYPVSTTNP*PNYSSTLSPSHSS 84
Score = 31.2 bits (69), Expect = 1.0
Identities = 38/137 (27%), Positives = 56/137 (40%), Gaps = 7/137 (5%)
Frame = -1
Query: 301 FSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLH 360
F S+SY + P N PS S + + +S++S Q + P S+P P S
Sbjct: 458 FLHLFSFSYSSFSPLQPLHNA-PPSTIPSPTFSSLSSSSHQQF-PRPSIPTIPISASSSS 285
Query: 361 KKNVNFDEYYPSPNFSPSPSPSSSSPIYS-----LGSLASKTLLRSESAPVNI-PNAEVT 414
+ + Y+P + S P S+P ++ L LAS + N PN T
Sbjct: 284 PFSSHSQSYHPLIHSSHFSLPFQSNPSHNPSPSYLHQLASNPQPHYPVSTTNP*PNYSST 105
Query: 415 YSPGHTSR-QNLPPSTP 430
SP H+S P TP
Sbjct: 104 LSPSHSSH*PYQTPETP 54
>TC90952 similar to GP|15724316|gb|AAL06551.1 AT4g34150/F28A23_90
{Arabidopsis thaliana}, partial (49%)
Length = 1046
Score = 40.8 bits (94), Expect = 0.001
Identities = 50/150 (33%), Positives = 65/150 (43%)
Frame = +2
Query: 281 FPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASS 340
+ ++P +PS+ PS P + HS +Y + + PAV Y P P + SH N SS
Sbjct: 395 YTAMPTPAVPSYNHVPSYYP-QKPHS-NYHH--PTTPAVAYPQPPP-YPASH----NPSS 547
Query: 341 QGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLR 400
PPSS P+P +S SP P PSSSSP S S+ S
Sbjct: 548 Y---PPSSTYPYPQSVS------------------SPYPPPSSSSPYPS--SVPSPYPPT 658
Query: 401 SESAPVNIPNAEVTYSPGHTSRQNLPPSTP 430
S S+P P +Y P S PPS P
Sbjct: 659 SSSSPSPYPPPH-SYPP---SSAYPPPSYP 736
>TC88399 weakly similar to GP|18491217|gb|AAL69511.1 putative AMP-binding
protein {Arabidopsis thaliana}, partial (62%)
Length = 1286
Score = 39.3 bits (90), Expect = 0.004
Identities = 35/117 (29%), Positives = 47/117 (39%)
Frame = +1
Query: 255 SEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRA 314
S TTP S + P D P P + SSPSSLP
Sbjct: 94 SSTTTPFSHGL------KPTNDASNLPPPSPP*VSAAATSSPSSLP-------------T 216
Query: 315 SPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYP 371
SPP +N SPSP+ S T + A ++ P SS +P+ S + + F + YP
Sbjct: 217 SPPCMNSTSPSPSPELSLTTSTRALTRVSSPTSSSTANPNSFSWISPHVILFSKLYP 387
>BQ751144 weakly similar to PIR|PQ0479|PQ04 pistil extensin-like protein
(clone pMG14) - common tobacco (fragment), partial (11%)
Length = 632
Score = 38.9 bits (89), Expect = 0.005
Identities = 43/147 (29%), Positives = 58/147 (39%), Gaps = 4/147 (2%)
Frame = +1
Query: 244 VMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSR 303
V R +S SS P+T SP + +P P P PS + PSSL FS
Sbjct: 160 VTRRRLLSLPSSPPSTSPSPHALLRSPTAPPAA-----PLSPPASSPSSPTRPSSL-FS- 318
Query: 304 RHSWSYENCRASPPAVNYYSPSPTHSESHTL----VSNASSQGYCPPSSLPPHPSEMSLL 359
PP+ + P P + S L + A+ PPSS PP P+ L
Sbjct: 319 ----------PPPPSPSRSRPPPPSTPSPLLPLPLLPLATEPSPFPPSSPPPRPASSPSL 468
Query: 360 HKKNVNFDEYYPSPNFSPSPSPSSSSP 386
+ F P +SPS PS+ +P
Sbjct: 469 SPPVLLFPPSASPPPWSPSRPPSALAP 549
Score = 34.3 bits (77), Expect = 0.12
Identities = 42/140 (30%), Positives = 59/140 (42%), Gaps = 12/140 (8%)
Frame = +1
Query: 290 PSHGSSPSSLPFSRRHSWSYENCR-------ASPPAVNYYSPSPTHSESHTLVSNASSQG 342
PS S+P S SW R +SPP+ SPSP H L+ + ++
Sbjct: 97 PSK*STPLPFLLSPVPSWPTVVTRRRLLSLPSSPPST---SPSP-----HALLRSPTAPP 252
Query: 343 YCP---PSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLG--SLASKT 397
P P+S P P+ S L F PSP+ S P PS+ SP+ L LA++
Sbjct: 253 AAPLSPPASSPSSPTRPSSL------FSPPPPSPSRSRPPPPSTPSPLLPLPLLPLATEP 414
Query: 398 LLRSESAPVNIPNAEVTYSP 417
S+P P + + SP
Sbjct: 415 SPFPPSSPPPRPASSPSLSP 474
>TC77148 ENOD20
Length = 1108
Score = 38.9 bits (89), Expect = 0.005
Identities = 36/110 (32%), Positives = 48/110 (42%), Gaps = 6/110 (5%)
Frame = +3
Query: 315 SPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLP-PHPSEMSLLHKKNVNFDEYYPSP 373
SPP +P P H +L S S PS P P P + H + + PSP
Sbjct: 441 SPPTPRSSTPIP-HPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASPSPSP 617
Query: 374 NFSPSPSPSSS-----SPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPG 418
+ S SPSPS S SP S+ SLA + S+ +P P+ + S G
Sbjct: 618 SLSKSPSPSESPSLAPSPSDSVASLAPSS-SPSDESPSPAPSPSSSGSKG 764
Score = 36.2 bits (82), Expect = 0.032
Identities = 46/147 (31%), Positives = 57/147 (38%), Gaps = 1/147 (0%)
Frame = +3
Query: 239 KLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSS 298
KL + VM P +S P+ P +P+ T P R + P P PS SPS
Sbjct: 384 KLAVVVMVAPVLSSPPPPPSPP-TPRSSTPIPHPPR--RSLPSPPSPSPS-PSPSPSPSP 551
Query: 299 LPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLV-SNASSQGYCPPSSLPPHPSEMS 357
P S + ASP S SP+ SES +L S + S PSS P S
Sbjct: 552 SPRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESP-- 725
Query: 358 LLHKKNVNFDEYYPSPNFSPSPSPSSS 384
SP+PSPSSS
Sbjct: 726 ------------------SPAPSPSSS 752
>TC84952 similar to PIR|T51759|T51759 [glutamate--ammonia-ligase]
adenylyltransferase (EC 2.7.7.42) [imported] -
Streptomyces coelicolor, partial (2%)
Length = 544
Score = 37.7 bits (86), Expect = 0.011
Identities = 19/48 (39%), Positives = 28/48 (57%)
Frame = +3
Query: 170 KKSTLLLRSLYATVRLLPAFKIFKELSSSANVSAFSLAHRVSTFFEPF 217
KK T+L S+ +LL +F I+ SS ++ FS+ +S FFEPF
Sbjct: 69 KKETILSLSIILNYKLLKSFNIWM*FLSSRKITNFSILSIISKFFEPF 212
>TC93234 weakly similar to GP|21751020|dbj|BAC03887. unnamed protein product
{Homo sapiens}, partial (11%)
Length = 591
Score = 37.4 bits (85), Expect = 0.015
Identities = 40/137 (29%), Positives = 52/137 (37%), Gaps = 4/137 (2%)
Frame = +2
Query: 254 SSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCR 313
SSE TP S + + R R PS P H S+ ++ PF+
Sbjct: 140 SSERATPASASLTSSSSIPSQRSRTSRSPS----SRPRHTSTGTAPPFA----------- 274
Query: 314 ASPPAVNYYSPSPTHSE----SHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEY 369
S PA + +PS T S S + S PPS+ PP P
Sbjct: 275 VSSPASSPTTPSSTRSSPGSTSTPRTPSTSFSARPPPSTPPPTPFRSR------------ 418
Query: 370 YPSPNFSPSPSPSSSSP 386
P+ SPS +PSSSSP
Sbjct: 419 -PTKGSSPSSTPSSSSP 466
>BI263340 weakly similar to PIR|T49223|T492 uclacyanin 3 [imported] -
Arabidopsis thaliana, partial (14%)
Length = 681
Score = 36.6 bits (83), Expect = 0.025
Identities = 49/164 (29%), Positives = 63/164 (37%), Gaps = 27/164 (16%)
Frame = +3
Query: 290 PSHGSSP-----SSLPFSRRHSWSY-------ENCRASPPAVNYYSPS---PTHSESHTL 334
PSHGSS SS P S H Y C SPP + PS P+H+++ +
Sbjct: 177 PSHGSSSPPSHGSSSPPS--HGGGYYTPTPPSTGCGYSPP----HDPSTSTPSHNQTPST 338
Query: 335 VSNASSQG--YCPPSSLPPHPSEMSLLHKKNVNFDE---YYPSPNFSPSPSPSSSS---- 385
SN S G Y P S P P + D P+P F PSPSP +
Sbjct: 339 PSNPPSSGGYYNSPPSTPTDPPVTLTPPSPSTPIDPGTPTVPTPPFLPSPSPFPGTCNYW 518
Query: 386 ---PIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLP 426
P G L + + NIP ++PG T Q+ P
Sbjct: 519 RTHPGIIWGILGWWGNMGNAFGVTNIP----XFNPGLTLPQHFP 638
>TC79359 weakly similar to PIR|S49915|S49915 extensin-like protein - maize,
partial (3%)
Length = 1170
Score = 35.8 bits (81), Expect = 0.042
Identities = 38/144 (26%), Positives = 58/144 (39%), Gaps = 8/144 (5%)
Frame = +3
Query: 345 PPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESA 404
PPSS+ P+ K+ +P SPS S S+ P S + + S +
Sbjct: 264 PPSSVSVSPTNSPASPAKSPTLSPPSQTPAVSPSGSASTPPPATSPPAKSPAVQPPSSVS 443
Query: 405 PVNIPNAEVTYSPGHTSRQNLPP-------STPIRISRCTSETE-RSRNLMQSCTTPEKM 456
P P+ V+ +P +S + PP S+P+ +S E S + S TP
Sbjct: 444 PAISPSNNVSSTPPVSSPASSPPTAAVSPVSSPVEAPSVSSPPEASSAGIPSSSATPADA 623
Query: 457 FSIGKESQKYSGGKIAPNSSPQIS 480
+ S K S G +SSP+ S
Sbjct: 624 PAATLPSSK-SPGTSPASSSPETS 692
>TC92805 similar to PIR|A86459|A86459 unknown protein 82634-81246
[imported] - Arabidopsis thaliana, partial (18%)
Length = 798
Score = 35.8 bits (81), Expect = 0.042
Identities = 30/122 (24%), Positives = 52/122 (42%), Gaps = 4/122 (3%)
Frame = -3
Query: 301 FSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLP----PHPSEM 356
F +H ++ + SPP+ + SP P+HS + S P SLP +
Sbjct: 793 FQTQHPSLSQS*QLSPPSSSSQSP-PSHSSRLSSNSEGGKSATKPKPSLPVLLFTSTTFT 617
Query: 357 SLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYS 416
S L + ++P P + +P+PS+ + +YS ++S L S NI + +
Sbjct: 616 SSLAFFPIPIPSFHPYPPSNAAPNPSTGTSLYSSNPISSGNLSISSLNFCNIETSTEAFG 437
Query: 417 PG 418
G
Sbjct: 436 LG 431
>TC82207 similar to GP|14517356|gb|AAK62569.1 AT4g15540/dl3810w {Arabidopsis
thaliana}, partial (50%)
Length = 990
Score = 35.8 bits (81), Expect = 0.042
Identities = 40/148 (27%), Positives = 60/148 (40%), Gaps = 4/148 (2%)
Frame = +1
Query: 308 SYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFD 367
S + P AV+ + HS+S T ++SQ +SLPP PS M N
Sbjct: 481 SLQEDEEKPGAVSPNIAAMLHSQSST---TSTSQLGDEDASLPPRPSSMQTNTSDAGNSY 651
Query: 368 EYYPSPNFSPSPSPSSSSPIYSLGS--LASKTLLRSESAPVNIPNAEVTYSPGHTSRQNL 425
+ P P P S S + LAS+T S P + P + SP TS+
Sbjct: 652 AEDRQSDAGPGPGPVRSQASSSSHNILLASQTTTPRISPPGSPPIVSASVSPSRTSKPAS 831
Query: 426 P--PSTPIRISRCTSETERSRNLMQSCT 451
P + ++I R +S + ++ S T
Sbjct: 832 PRRHAVSLQIDRTSSMFSSTGSMSSSGT 915
>TC86493 similar to GP|18654317|gb|AAL77589.1 ribose-5-phosphate isomerase
{Spinacia oleracea}, partial (80%)
Length = 1109
Score = 35.4 bits (80), Expect = 0.055
Identities = 34/127 (26%), Positives = 50/127 (38%), Gaps = 18/127 (14%)
Frame = +1
Query: 322 YSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSP 381
+ P P+H H+L P SS+PPH + L P N++P+PSP
Sbjct: 91 WHPYPSHHHHHSL----------PHSSMPPHDLTYAPL-----------PLLNYAPNPSP 207
Query: 382 SSSSPIYSLGSLA----------------SKTLLRSESAPVNIPNAEVTYSPGH--TSRQ 423
S S P SL + + S L + P + PN+ +P + TS
Sbjct: 208 SPSKP*PSLKTTSKNSPPIKPSNMSKAAWSSALALAPPPPSSSPNSANFSTPANSPTSLA 387
Query: 424 NLPPSTP 430
PP+ P
Sbjct: 388 FPPPNAP 408
>BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (30%)
Length = 1338
Score = 35.4 bits (80), Expect = 0.055
Identities = 29/121 (23%), Positives = 45/121 (36%), Gaps = 1/121 (0%)
Frame = -1
Query: 296 PSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSL-PPHPS 354
P + ++ S+ ++PPA++++ P TL S CPP S PPHP
Sbjct: 642 PGRVLLVMQYPTSFLGTGSAPPAIHHHPNPPASQSPATLNRPTSHTPACPPPSRPPPHPR 463
Query: 355 EMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVT 414
+P+ SP P P + G S TL + P+A T
Sbjct: 462 -----------------APSLSPPPPPHPPTSPPRPGRPRSSTLAPPPHRQFHSPSALCT 334
Query: 415 Y 415
+
Sbjct: 333 H 331
Score = 32.7 bits (73), Expect = 0.36
Identities = 32/108 (29%), Positives = 44/108 (40%), Gaps = 6/108 (5%)
Frame = -1
Query: 285 PVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYC 344
P R P H +PS P H + R P + +P P H + H+ + + G
Sbjct: 492 PPSRPPPHPRAPSLSPPPPPHPPTSPP-RPGRPRSSTLAPPP-HRQFHSPSALCTHCGIP 319
Query: 345 PPSSLPPHP------SEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSP 386
PP+ PP P S +L+ F +P P PSPSPS P
Sbjct: 318 PPAVPPPPPPTPPPSSPPALVSPPPRPF--LFPPPPNQPSPSPSPPPP 181
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.127 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,351,460
Number of Sequences: 36976
Number of extensions: 365739
Number of successful extensions: 3997
Number of sequences better than 10.0: 211
Number of HSP's better than 10.0 without gapping: 2993
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3542
length of query: 535
length of database: 9,014,727
effective HSP length: 101
effective length of query: 434
effective length of database: 5,280,151
effective search space: 2291585534
effective search space used: 2291585534
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146842.6


