

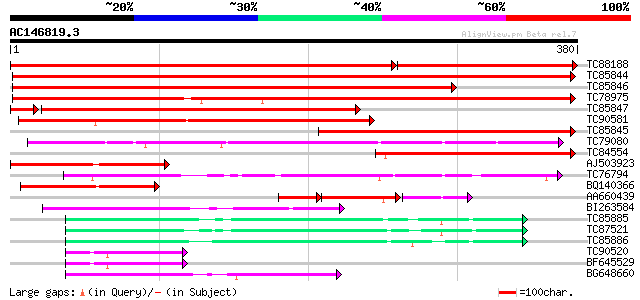
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146819.3 + phase: 0
(380 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88188 homologue to SP|P80572|ADHX_PEA Alcohol dehydrogenase cl... 534 0.0
TC85844 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 ... 476 e-135
TC85846 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 ... 407 e-114
TC78975 similar to GP|21592656|gb|AAM64605.1 alcohol dehydrogena... 402 e-112
TC85847 homologue to SP|P13603|ADH1_TRIRP Alcohol dehydrogenase ... 286 2e-80
TC90581 similar to PIR|E86357|E86357 alcohol dehydrogenase homol... 212 2e-55
TC85845 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 ... 197 4e-51
TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogena... 164 7e-41
TC84554 weakly similar to GP|7705214|gb|AAB33480.2| alcohol dehy... 125 4e-29
AJ503923 similar to GP|10129655|emb alcohol dehydrogenase-like p... 116 1e-26
TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbi... 103 1e-22
BQ140366 weakly similar to SP|P41681|ADH2_ Alcohol dehydrogenase... 91 6e-19
AA660439 similar to PIR|S52036|S520 probable alcohol dehydrogena... 60 8e-19
BI263584 similar to SP|Q9P6C8|ADH1 Alcohol dehydrogenase I (EC 1... 76 2e-14
TC85885 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 73 2e-13
TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 73 2e-13
TC85886 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 70 1e-12
TC90520 weakly similar to PIR|T08581|T08581 cinnamyl-alcohol deh... 66 2e-11
BF645529 weakly similar to GP|22475166|gb putative sinapyl alcoh... 66 2e-11
BG648660 similar to SP|Q9ZRF1|MTD_ Probable mannitol dehydrogena... 61 6e-10
>TC88188 homologue to SP|P80572|ADHX_PEA Alcohol dehydrogenase class III (EC
1.1.1.1) (Glutathione-dependent formaldehyde
dehydrogenase), complete
Length = 1597
Score = 534 bits (1375), Expect(2) = 0.0
Identities = 259/259 (100%), Positives = 259/259 (100%)
Frame = +1
Query: 1 MASSTEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKD 60
MASSTEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKD
Sbjct: 223 MASSTEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKD 402
Query: 61 PEGLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRA 120
PEGLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRA
Sbjct: 403 PEGLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRA 582
Query: 121 ATGVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCG 180
ATGVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCG
Sbjct: 583 ATGVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCG 762
Query: 181 VPTGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNF 240
VPTGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNF
Sbjct: 763 VPTGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNF 942
Query: 241 GVTEFINPKDHEKPIQQVI 259
GVTEFINPKDHEKPIQQVI
Sbjct: 943 GVTEFINPKDHEKPIQQVI 999
Score = 249 bits (637), Expect(2) = 0.0
Identities = 120/120 (100%), Positives = 120/120 (100%)
Frame = +2
Query: 261 DLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQEISTRPFQLVTGRVWK 320
DLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQEISTRPFQLVTGRVWK
Sbjct: 1004 DLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQEISTRPFQLVTGRVWK 1183
Query: 321 GTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAFDLMHEGKCLRCVLALH 380
GTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAFDLMHEGKCLRCVLALH
Sbjct: 1184 GTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAFDLMHEGKCLRCVLALH 1363
>TC85844 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 (EC
1.1.1.1). [Garden pea] {Pisum sativum}, complete
Length = 1502
Score = 476 bits (1226), Expect = e-135
Identities = 218/377 (57%), Positives = 288/377 (75%)
Frame = +2
Query: 3 SSTEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPE 62
S+T GQ+I C+AAVAWE KPL +E+VEVAPPQA EVR++ILFT+LCHTD Y W K
Sbjct: 92 SNTAGQIIKCRAAVAWEAGKPLVMEEVEVAPPQAGEVRLKILFTSLCHTDVYFWEAKGQT 271
Query: 63 GLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAAT 122
LFP I GHEA GIVES+GEGVT +KPGDH +P + ECG+C CKS ++N+C +R T
Sbjct: 272 PLFPRIFGHEAGGIVESIGEGVTHLKPGDHALPVFTGECGDCPHCKSEESNMCDLLRINT 451
Query: 123 GVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVP 182
GVM+ND + RFS+KG+PI+HF+GTSTFS+YTVVH VAKI+PDA LDKVC+L CG+
Sbjct: 452 DRGVMINDNQSRFSLKGQPIHHFVGTSTFSEYTVVHAGCVAKINPDAPLDKVCILSCGIC 631
Query: 183 TGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGV 242
TGLGA N AK +PGS VAIFGLG VGLA AEGA+ +GASRIIG+D+ S++F+ AK FGV
Sbjct: 632 TGLGATINVAKPKPGSSVAIFGLGAVGLAAAEGARISGASRIIGVDLVSSRFELAKKFGV 811
Query: 243 TEFINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAAS 302
EF+NPKDH+KP+QQVI+++T+GGVD + EC G++ M SA EC H GWG +V+VGV
Sbjct: 812 NEFVNPKDHDKPVQQVIAEMTNGGVDRAVECTGSIQAMISAFECVHDGWGVAVLVGVPNK 991
Query: 303 GQEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINE 362
T P L+ R KGT +G +K R+ +P +VEKY+K E+++++++TH + +EIN+
Sbjct: 992 DDAFKTHPMNLLNERTLKGTFYGNYKPRTDLPNVVEKYMKGELELEKFITHTIPFSEINK 1171
Query: 363 AFDLMHEGKCLRCVLAL 379
AFD M +G+ +RC++ +
Sbjct: 1172AFDYMLKGESIRCIIRM 1222
>TC85846 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 (EC
1.1.1.1). [Garden pea] {Pisum sativum}, partial (78%)
Length = 955
Score = 407 bits (1046), Expect = e-114
Identities = 191/297 (64%), Positives = 235/297 (78%)
Frame = +3
Query: 3 SSTEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPE 62
S+T QVI C+AAVAWE KPL +E+VEVAPPQA EVR++ILFT+LCHTD Y W K
Sbjct: 48 SNTTVQVIKCRAAVAWEAGKPLVMEEVEVAPPQAGEVRLKILFTSLCHTDVYFWEAKGQT 227
Query: 63 GLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAAT 122
LFP I GHEA GIVESVGEGVT +KPGDH +P + ECG+C CKS ++N+C +R T
Sbjct: 228 PLFPRIFGHEAGGIVESVGEGVTHLKPGDHALPVFTGECGDCPHCKSEESNMCDLLRINT 407
Query: 123 GVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVP 182
GVM+ND + RFSIKG+PI+HF+GTSTFS+YTVVH VAKI+PDA LDKVC+L CG+
Sbjct: 408 DRGVMINDNQSRFSIKGQPIHHFVGTSTFSEYTVVHAGCVAKINPDAPLDKVCILSCGIC 587
Query: 183 TGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGV 242
TGLGA N AK +PGS VAIFGLG VGLA AEGA+ +GASRIIG+D+ S++F+ AK FGV
Sbjct: 588 TGLGATINVAKPKPGSSVAIFGLGAVGLAAAEGARISGASRIIGVDLVSSRFELAKKFGV 767
Query: 243 TEFINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGV 299
EF+NPKDH+KP+QQVI+++T+GGVD + EC G++ M SA EC H GWG +V+VGV
Sbjct: 768 NEFVNPKDHDKPVQQVIAEMTNGGVDRAVECTGSIQAMISAFECVHDGWGVAVLVGV 938
>TC78975 similar to GP|21592656|gb|AAM64605.1 alcohol dehydrogenase
putative {Arabidopsis thaliana}, partial (77%)
Length = 1691
Score = 402 bits (1032), Expect = e-112
Identities = 203/385 (52%), Positives = 270/385 (69%), Gaps = 8/385 (2%)
Frame = +2
Query: 3 SSTEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGK-DP 61
S T+G+ ITCKAAVA P +P +E + V PPQ EVRI+IL+T++CHTD W G+ +P
Sbjct: 104 SGTKGKTITCKAAVAHGPGEPFVVEQILVQPPQKMEVRIKILYTSICHTDLSGWKGECEP 283
Query: 62 EGLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAA 121
+ +P I GHEA+GIVESVGEGV D+K D V+ + ECGECK C KTN+C K
Sbjct: 284 QRAYPRIFGHEASGIVESVGEGVNDMKENDKVVLIFNGECGECKCCMCKKTNMCEK---- 451
Query: 122 TGVGVM---MNDRKPRFS-IKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPD--AALDKVC 175
+GV M M D RFS I GKPI+HF+ TSTF++YTVV K++ + +L K+
Sbjct: 452 SGVDPMKKLMCDGTSRFSSIDGKPIFHFLNTSTFTEYTVVDSACAVKLNTEDNLSLKKLT 631
Query: 176 LLGCGVPTGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFD 235
LL CGV TG+GA WN A V GS VAIFGLG VGLAVAEGA++ GAS+IIG+DI+ +KF+
Sbjct: 632 LLSCGVSTGIGAAWNNANVHAGSSVAIFGLGAVGLAVAEGARARGASKIIGVDINPDKFN 811
Query: 236 -TAKNFGVTEFINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTS 294
TA+ G+TEFINPKD EKP+ ++I ++TDGGVDYSFEC GN++V+R + H+GWG +
Sbjct: 812 NTAETMGITEFINPKDEEKPVYEIIREMTDGGVDYSFECTGNLNVLRDSFLSVHEGWGLT 991
Query: 295 VVVGVAASGQEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHN 354
V++G+ S + + P +L GR +G+ FGGFK +SQ+P L + +K IK+D ++TH
Sbjct: 992 VLLGIHGSPKLLPIHPMELFDGRRIEGSVFGGFKGKSQLPNLATECMKGAIKLDNFITHE 1171
Query: 355 LTLAEINEAFDLMHEGKCLRCVLAL 379
L EIN+AF+L+ GK LRCVL L
Sbjct: 1172LPFDEINQAFNLLIAGKSLRCVLTL 1246
>TC85847 homologue to SP|P13603|ADH1_TRIRP Alcohol dehydrogenase 1 (EC
1.1.1.1). [Creeping white clover] {Trifolium repens},
partial (60%)
Length = 751
Score = 286 bits (731), Expect(2) = 2e-80
Identities = 137/214 (64%), Positives = 167/214 (78%)
Frame = +1
Query: 22 KPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGLFPCILGHEAAGIVESVG 81
KPL +E+VEVAPPQA EVR++ILFT+LCHTD Y W K LFP I GHEA GIVESVG
Sbjct: 109 KPLVMEEVEVAPPQAGEVRLKILFTSLCHTDVYFWEAKGQTPLFPRIFGHEAGGIVESVG 288
Query: 82 EGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGVGVMMNDRKPRFSIKGKP 141
EGVT + PGDH + + ECG+C C S ++N+C +R T GVM+ND + RFSIKG+P
Sbjct: 289 EGVTHLTPGDHALLVFTGECGDCPHCMSEESNMCDLLRINTDRGVMINDNQSRFSIKGQP 468
Query: 142 IYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPTGLGAVWNTAKVEPGSIVA 201
I+HF+GTSTFS+YTVVH VAKI+PDA LDKVC+L CG+ TGLGA N AK +PGS VA
Sbjct: 469 IHHFVGTSTFSEYTVVHAGCVAKINPDAPLDKVCILSCGICTGLGATINVAKPKPGSSVA 648
Query: 202 IFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFD 235
IFGLG VGLA AEGA+ +GASRIIG+D+ S++F+
Sbjct: 649 IFGLGAVGLAAAEGARISGASRIIGVDLVSSRFE 750
Score = 31.6 bits (70), Expect(2) = 2e-80
Identities = 13/19 (68%), Positives = 16/19 (83%)
Frame = +3
Query: 1 MASSTEGQVITCKAAVAWE 19
M+++T GQVI C AAVAWE
Sbjct: 45 MSNTTTGQVIKCTAAVAWE 101
>TC90581 similar to PIR|E86357|E86357 alcohol dehydrogenase homolog
[imported] - Arabidopsis thaliana, partial (52%)
Length = 1289
Score = 212 bits (540), Expect = 2e-55
Identities = 118/243 (48%), Positives = 154/243 (62%), Gaps = 5/243 (2%)
Frame = +1
Query: 7 GQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTW--SGKDPEGL 64
G+ I CKAA+ + +PL IE+VE+ PP++ EVRI+IL T+LCH+D W + P
Sbjct: 277 GKPIRCKAAICKKSGEPLVIEEVELDPPKSWEVRIKILCTSLCHSDVTFWKMNSSAPTAR 456
Query: 65 FPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGV 124
FP ILGHEA G+VESVGE V +VK GD V+P + CGEC C S K+N C K +
Sbjct: 457 FPRILGHEAVGLVESVGENVEEVKEGDLVVPVFLPNCGECIDCGSTKSNNCTKF-GNKPI 633
Query: 125 GVMMNDRKPRF-SIKGKPIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPT 183
M D RF +KG+ ++H +G S+FS+YTVV V KI D LDK CLL CGV T
Sbjct: 634 RDMPRDGTSRFRDMKGEVVHHLLGVSSFSEYTVVDVTHVVKITHDIPLDKACLLSCGVST 813
Query: 184 GLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRII-GIDIDSNKFDTAKN-FG 241
G+GA W A VE G+ VAIFGLG VGLAVA K GAS+I +D++ ++ +N G
Sbjct: 814 GIGAAWKVADVEKGTTVAIFGLGAVGLAVAVATKQRGASKIYRRVDLNHDEI*NEENKCG 993
Query: 242 VTE 244
+T+
Sbjct: 994 ITD 1002
>TC85845 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 (EC
1.1.1.1). [Garden pea] {Pisum sativum}, partial (45%)
Length = 734
Score = 197 bits (502), Expect = 4e-51
Identities = 86/172 (50%), Positives = 128/172 (74%)
Frame = +1
Query: 208 VGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTEFINPKDHEKPIQQVISDLTDGGV 267
VGLA AEGA+ +GASRIIG+D+ S++F+ AK FGV EF+NPKDH+KP+QQVI+++T+GGV
Sbjct: 1 VGLAAAEGARISGASRIIGVDLVSSRFELAKKFGVNEFVNPKDHDKPVQQVIAEMTNGGV 180
Query: 268 DYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQEISTRPFQLVTGRVWKGTAFGGF 327
D + EC G++ M SA EC H GWG +V+VGV T P L+ R KGT +G +
Sbjct: 181 DRAVECTGSIQAMISAFECVHDGWGVAVLVGVPNKDDAFQTHPMNLLNERTLKGTFYGNY 360
Query: 328 KSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAFDLMHEGKCLRCVLAL 379
K R+ +P +VEKY+K E+++++++TH + +EIN+AFD M +G+ +RC++ +
Sbjct: 361 KPRTDLPNVVEKYMKGELELEKFITHTIPFSEINKAFDYMLKGESIRCIIRM 516
>TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogenase-like
protein {Arabidopsis thaliana}, partial (96%)
Length = 1524
Score = 164 bits (414), Expect = 7e-41
Identities = 112/367 (30%), Positives = 177/367 (47%), Gaps = 8/367 (2%)
Frame = +3
Query: 13 KAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGLFPCILGHE 72
+ AV EPNKPLTIE+ + P+A E+ I+ +CH+D + G+ P PC++GHE
Sbjct: 234 RGAVFREPNKPLTIEEFHIPRPKAGELLIKTKGCGVCHSDLHVMKGEIPFSS-PCVVGHE 410
Query: 73 AAGIVESVGEGVTDVKP------GDHVIPCYQAECGECKFCKSGKTNLCGKVRAATGVGV 126
G V G+ TD K G V+ + CG C +C G +LC A
Sbjct: 411 ITGEVVEHGQH-TDSKTIERLPIGSRVVGAFIMPCGNCSYCSKGHDDLCEAFFAYNRAKG 587
Query: 127 MMNDRKPRFSIKGK--PIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGCGVPTG 184
+ D + R ++G PIY + ++Y VV ++A + + +LGC V T
Sbjct: 588 TLYDGETRLFLRGSGNPIYMY-SMGGLAEYCVVPANALAVLPNSMPYTESAILGCAVFTA 764
Query: 185 LGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTE 244
GA+ + A+V PG VA+ G G VG + + A++ GAS II +D+ K + AK G T
Sbjct: 765 YGAMAHAAEVRPGDTVAVIGTGGVGSSCLQIARAFGASDIIAVDVQDEKLEKAKTLGATH 944
Query: 245 FINPKDHEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQ 304
IN E PI++++ GVD + E +G + G G +V++G+A +G
Sbjct: 945 TIN-SAKEDPIEKILEITGGKGVDVAVEALGRPQTFAQCTQSVKDG-GKAVMIGLAQAGS 1118
Query: 305 EISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEAF 364
+LV ++ ++GG ++R +P L+ + VT T E +AF
Sbjct: 1119LGEVDINRLVRRKIKVIGSYGG-RARQDLPKLIRLAETGIFDLGHAVTRKYTFEESGKAF 1295
Query: 365 DLMHEGK 371
++EGK
Sbjct: 1296QDLNEGK 1316
>TC84554 weakly similar to GP|7705214|gb|AAB33480.2| alcohol dehydrogenase
ADH {Lycopersicon esculentum}, partial (27%)
Length = 550
Score = 125 bits (313), Expect = 4e-29
Identities = 52/136 (38%), Positives = 88/136 (64%), Gaps = 2/136 (1%)
Frame = +2
Query: 246 INPKD--HEKPIQQVISDLTDGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASG 303
+NP +EK + +VI D+T+GG DY FECIG S+M A + +GWG +V++GV G
Sbjct: 2 VNPSSTSNEKSVSEVIKDMTNGGADYCFECIGLASLMTEAFNSSREGWGKTVIIGVEMHG 181
Query: 304 QEISTRPFQLVTGRVWKGTAFGGFKSRSQVPWLVEKYLKKEIKVDEYVTHNLTLAEINEA 363
++ P+ ++ G+ G+ FGG K +S +P L +KYL KE+ +D +++ + +IN+A
Sbjct: 182 SPLTLNPYDILKGKTITGSLFGGLKPKSDLPLLAQKYLDKELNLDGFISQEVDFKDINKA 361
Query: 364 FDLMHEGKCLRCVLAL 379
FD + +GK +RC++ +
Sbjct: 362 FDYLLQGKSIRCIIRM 409
>AJ503923 similar to GP|10129655|emb alcohol dehydrogenase-like protein
{Arabidopsis thaliana}, partial (25%)
Length = 350
Score = 116 bits (291), Expect = 1e-26
Identities = 57/107 (53%), Positives = 74/107 (68%)
Frame = +2
Query: 1 MASSTEGQVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKD 60
MASS+ QVITCKAAVAW + L +E+VEV+PPQ E+RI+++ T+LC +D W +
Sbjct: 38 MASSSTPQVITCKAAVAWGAGEALVMEEVEVSPPQPLEIRIKVVSTSLCRSDLSAW---E 208
Query: 61 PEGLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAECGECKFC 107
+FP I GHEA+GIVESVG GVT+ K GD V+ + EC CK C
Sbjct: 209 THAIFPRISGHEASGIVESVGLGVTEFKEGDQVLTVFIGECMSCKLC 349
>TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbitol
dehydrogenase {Prunus persica}, partial (93%)
Length = 1691
Score = 103 bits (256), Expect = 1e-22
Identities = 92/345 (26%), Positives = 149/345 (42%), Gaps = 11/345 (3%)
Frame = +2
Query: 37 NEVRIQILFTALCHTDAY---TWSGKDPEGLFPCILGHEAAGIVESVGEGVTDVKPGDHV 93
++VRI++ +C +D + T D P ++GHE AGI+E VG V + PGD V
Sbjct: 332 HDVRIKMKAVGICGSDVHYLKTLRCADFIVKEPMVIGHECAGIIEEVGSQVKTLVPGDRV 511
Query: 94 IPCYQAECGECKFCKSGKTNLCGKVRAATGVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQ 153
C C CK G+ NLC P P H S +Q
Sbjct: 512 AIEPGISCWRCDHCKLGRYNLC-----------------PDMKFFATPPVH---GSLANQ 631
Query: 154 YTVVHDVSVA-KIHPDAALDKVCLLGCGVPTGLGA-VWNTAKVEPGSIVAIFGLGTVGLA 211
+VH + K+ + +L++ + P +G A + P + V I G G +GL
Sbjct: 632 --IVHPADLCFKLPENVSLEEGAMCE---PLSVGVHACRRANIGPETNVLIMGAGPIGLV 796
Query: 212 VAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTEFI----NPKDHEKPIQQVISDLTDGGV 267
A++ GA RI+ +D+D ++ AK+ G + + N +D + ++Q I ++ GV
Sbjct: 797 TMLSARAFGAPRIVVVDVDDHRLSVAKSLGADDIVKVSTNIQDVAEEVKQ-IHNVLGAGV 973
Query: 268 DYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQEISTRPFQLVTGRVWKGTAFGGF 327
D +F+C G M +AL T G G +VG+ S + P + G F
Sbjct: 974 DVTFDCAGFNKTMTTALTATQPG-GKVCLVGMGHSEMTVPLTP-----AAAREVDVVGIF 1135
Query: 328 KSRSQVPWLVEKYLKKEIKVDEYVTHNLTLA--EINEAFDLMHEG 370
+ ++ P +E +I V +TH + E+ EAF+ G
Sbjct: 1136RYKNTWPLCLEFLRSGKIDVKPLITHRFGFSQKEVEEAFETSARG 1270
>BQ140366 weakly similar to SP|P41681|ADH2_ Alcohol dehydrogenase 2 (EC
1.1.1.1). [Deer mouse] {Peromyscus maniculatus}, partial
(11%)
Length = 327
Score = 91.3 bits (225), Expect = 6e-19
Identities = 41/93 (44%), Positives = 63/93 (67%)
Frame = +3
Query: 8 QVITCKAAVAWEPNKPLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDPEGLFPC 67
+++ K AV W KP+T+E+++V PP+A EVR+++L +++CHTD + G P FP
Sbjct: 51 RLLHAKPAVCWGLGKPVTVEEIQVDPPKAKEVRVKMLCSSVCHTDISSLKG-FPHNQFPL 227
Query: 68 ILGHEAAGIVESVGEGVTDVKPGDHVIPCYQAE 100
LGHE G+VES+G+ VT++K GD + P Y E
Sbjct: 228 XLGHEGIGVVESIGDQVTNLKEGDFIXPTYIGE 326
>AA660439 similar to PIR|S52036|S520 probable alcohol dehydrogenase (EC
1.1.1.1) ADH3b - tomato, partial (21%)
Length = 403
Score = 59.7 bits (143), Expect(3) = 8e-19
Identities = 27/55 (49%), Positives = 40/55 (72%), Gaps = 2/55 (3%)
Frame = +2
Query: 210 LAVAEGAKSAGASRIIGIDIDSNKFDTAKNFGVTEFINPK--DHEKPIQQVISDL 262
LAVA AK GAS+IIG+D++ +KF+ K FG+T+F+NP +EK + +VI D+
Sbjct: 89 LAVAVAAKQRGASKIIGVDLNHDKFEIGKQFGITDFVNPSSTSNEKSVSEVIKDM 253
Score = 42.4 bits (98), Expect(3) = 8e-19
Identities = 19/29 (65%), Positives = 21/29 (71%)
Frame = +3
Query: 181 VPTGLGAVWNTAKVEPGSIVAIFGLGTVG 209
V TG+GA W A VE G+ VAIFGLG VG
Sbjct: 3 VSTGIGAAWKVADVEKGTTVAIFGLGAVG 89
Score = 28.9 bits (63), Expect(3) = 8e-19
Identities = 14/47 (29%), Positives = 21/47 (43%)
Frame = +1
Query: 264 DGGVDYSFECIGNVSVMRSALECTHKGWGTSVVVGVAASGQEISTRP 310
+GG DY FECIG ++M L + G + +G + P
Sbjct: 256 NGGADYCFECIGLATLMTEXLIAVXR-LGXXXXIXXECTGSXFNLNP 393
>BI263584 similar to SP|Q9P6C8|ADH1 Alcohol dehydrogenase I (EC 1.1.1.1).
{Neurospora crassa}, partial (57%)
Length = 682
Score = 75.9 bits (185), Expect = 2e-14
Identities = 54/206 (26%), Positives = 93/206 (44%), Gaps = 4/206 (1%)
Frame = +2
Query: 23 PLTIEDVEVAPPQANEVRIQILFTALCHTDAYTWSGKDP-EGLFPCILGHEAAGIVESVG 81
P+ + + V P +EV + + F+ +CHTD + W G P + P + GHE AG+V + G
Sbjct: 122 PIEYKQIPVQKPGPDEVLVNVKFSGVCHTDLHAWQGDWPLDTKLPLVGGHEGAGVVVARG 301
Query: 82 EGVTDVKPGDHV-IPCYQAECGECKFCKSGKTNLCGKVRAATGVGVMMNDRKPRFSIKGK 140
+ V DVK G+ V I C C +C++ +LC + + +++ G
Sbjct: 302 DLVKDVKIGEKVGIKWLNGSCLSCSYCQNADESLCAEALLS------------GYTVDG- 442
Query: 141 PIYHFMGTSTFSQYTVVHDVSVAKIHPDAALDKVCLLGC-GVPTGLGAVWNTAKVEPGSI 199
+F QY + + VA+I + L+ + + C G+ G + V+ G
Sbjct: 443 ---------SFQQYAIAKAIHVARIPEECDLESISPILCAGITVYKGL--KESGVKAGQS 589
Query: 200 VAIFGL-GTVGLAVAEGAKSAGASRI 224
+AI G G +G + K+ G I
Sbjct: 590 IAIVGAGGGLGSIAXQYCKAMGIHAI 667
>TC85885 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (97%)
Length = 1330
Score = 73.2 bits (178), Expect = 2e-13
Identities = 79/315 (25%), Positives = 125/315 (39%), Gaps = 5/315 (1%)
Frame = +3
Query: 38 EVRIQILFTALCHTDAYTWSGKDPEGLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCY 97
+V ++L+ +CHTD + + ++P + GHE AGIV VG V K GD V Y
Sbjct: 183 DVAFKVLYCGICHTDLHMMKNEWGNSIYPLVPGHELAGIVTEVGSKVEKFKVGDKVGVGY 362
Query: 98 QAE-CGECKFCKSGKTNLCGKVRAATGVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTV 156
+ C C+ C N C + G G Y +S V
Sbjct: 363 MVDSCRSCENCAEDLENYCPQQTVTCGAKYR----------DGSVTY-----GGYSDSMV 497
Query: 157 VHDVSVAKIHPDAALDKVCLLGCGVPTGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGA 216
+ V +I LD L C T + + +PG + + GLG +G + A
Sbjct: 498 ADEHFVIRIPDSLPLDVAGPLLCAGVTVYSPLRHFQLDKPGMNIGVVGLGGLGHMAVKFA 677
Query: 217 KSAGASRIIGIDIDSNKFDTAKNFGVTEFINPKDHEKPIQQVISDLTDGGVDYSFECIGN 276
K+ GA+ + S + + ++ G F+ +D P Q + T G+ I
Sbjct: 678 KAFGANVTVISTSPSKEKEAIEHLGADSFLVSRD---PDQMQAAMGTLNGI------IDT 830
Query: 277 VSVMRSALECTH--KGWGTSVVVGVAASGQEISTRPFQLVTGR-VWKGTAFGGFK-SRSQ 332
VS L K G V+VG A E+ F L+ GR + G+ GG K ++
Sbjct: 831 VSASHPILPLIGLLKSNGKLVMVGGVAKPLELPV--FSLLGGRKLVAGSLIGGIKETQEM 1004
Query: 333 VPWLVEKYLKKEIKV 347
+ + E + +I+V
Sbjct: 1005IDFAAEHNVTPDIEV 1049
>TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (98%)
Length = 1433
Score = 72.8 bits (177), Expect = 2e-13
Identities = 78/315 (24%), Positives = 124/315 (38%), Gaps = 5/315 (1%)
Frame = +2
Query: 38 EVRIQILFTALCHTDAYTWSGKDPEGLFPCILGHEAAGIVESVGEGVTDVKPGDHV-IPC 96
+V ++L+ +CH+D + + +P + GHE AGIV VG V K GD V + C
Sbjct: 182 DVAFKVLYCGICHSDLHMIKNEWGMSTYPLVPGHEIAGIVTEVGSKVEKFKIGDKVGVGC 361
Query: 97 YQAECGECKFCKSGKTNLCGKVRAATGVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTV 156
C C+ C+ N C K N ++S G Y +S V
Sbjct: 362 LVDSCRACQNCEENLENYCPK---------QTNTYSAKYS-DGSITY-----GGYSDSMV 496
Query: 157 VHDVSVAKIHPDAALDKVCLLGCGVPTGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGA 216
+ + I L+ L C T + +PG + I GLG +G + A
Sbjct: 497 ADEHFIVHIPDGLPLESAAPLLCAGITVYSPLRYFGLDKPGMNIGIVGLGGLGHLGVKFA 676
Query: 217 KSAGASRIIGIDIDSNKFDTAKNFGVTEFINPKDHEKPIQQVISDLTDGGVDYSFECIGN 276
K+ GA+ + + + + +N G F+ D +K Q DG +D
Sbjct: 677 KAFGANVTVISTSPNKEKEAIENLGADSFLISHDQDK--MQAAMGTLDGIID-------T 829
Query: 277 VSVMRSALECTH--KGWGTSVVVGVAASGQEISTRPFQLVTGR-VWKGTAFGGFK-SRSQ 332
VS L K G V+VG E+ P L+ GR G+ GG K ++
Sbjct: 830 VSADHPLLPLVGLLKYHGKLVMVGAPDKPPELPHIP--LIMGRKTISGSGIGGMKETQEM 1003
Query: 333 VPWLVEKYLKKEIKV 347
+ + + +K +I+V
Sbjct: 1004IDFAAKHNIKPDIEV 1048
>TC85886 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (94%)
Length = 1395
Score = 70.1 bits (170), Expect = 1e-12
Identities = 73/316 (23%), Positives = 126/316 (39%), Gaps = 6/316 (1%)
Frame = +2
Query: 38 EVRIQILFTALCHTDAYTWSGKDPEGLFPCILGHEAAGIVESVGEGVTDVKPGDHVIPCY 97
+V ++L+ +CH+D + + + +P + GHE G+V VG V K GD V Y
Sbjct: 185 DVAFKVLYCGICHSDLHIMQNEWGKTTYPLVPGHELTGVVTEVGSKVKKFKVGDKVGVGY 364
Query: 98 QAE-CGECKFCKSGKTNLCGKVRAATGVGVMMNDRKPRFSIKGKPIYHFMGTSTFSQYTV 156
+ C C+ C N C K + GK + FS V
Sbjct: 365 MVDSCRSCENCADDIENYCTKYTQ---------------TFNGKSRDGTITYGGFSDSMV 499
Query: 157 VHDVSVAKIHPDAALDKVCLLGCGVPTGLGAVWNTAKVEPGSIVAIFGLGTVGLAVAEGA 216
+ V +I LD L C T + + +PG + + GLG +G + A
Sbjct: 500 ADEHFVIRIPDSLPLDGAGPLLCAGVTVYSPLRHFGLDKPGMNIGVVGLGGLGHMAVKFA 679
Query: 217 KSAGASRIIGIDIDSNKFDTAKNFGVTEFINPKDHEKPIQQVISDLTDGGVD---YSFEC 273
K+ GA + + + ++ G F+ +D E+ Q + +G +D S
Sbjct: 680 KAFGAKVTVISTSPKKEKEAIEHLGADSFLVSRDPEQ--MQAATSTLNGIIDTVSASHPV 853
Query: 274 IGNVSVMRSALECTHKGWGTSVVVGVAASGQEISTRPFQLVTGR-VWKGTAFGGFK-SRS 331
+ + +++S G V+VG A E+ F L+ GR G+ GG K ++
Sbjct: 854 VPLIGLLKSN--------GKLVMVGAVAKPLELPI--FSLLGGRKSIAGSLIGGIKETQE 1003
Query: 332 QVPWLVEKYLKKEIKV 347
+ + + + EI+V
Sbjct: 1004MIDFAAKHNVTPEIEV 1051
>TC90520 weakly similar to PIR|T08581|T08581 cinnamyl-alcohol dehydrogenase
(EC 1.1.1.195) CAD1 - Arabidopsis thaliana, partial
(41%)
Length = 618
Score = 66.2 bits (160), Expect = 2e-11
Identities = 32/85 (37%), Positives = 50/85 (58%), Gaps = 3/85 (3%)
Frame = +2
Query: 38 EVRIQILFTALCHTDAYTWSGKDPEGL--FPCILGHEAAGIVESVGEGVTDVKPGDHV-I 94
+V I+IL+ +CHTD + KD G+ +P + GHE G++ VG V K GD V +
Sbjct: 149 DVTIKILYCGICHTDVH--HAKDDWGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGV 322
Query: 95 PCYQAECGECKFCKSGKTNLCGKVR 119
C A C +C++CK+ + N C K++
Sbjct: 323 GCLAASCLDCEYCKTDQENYCEKLQ 397
>BF645529 weakly similar to GP|22475166|gb putative sinapyl alcohol
dehydrogenase {Populus tremula x Populus tremuloides},
partial (40%)
Length = 658
Score = 66.2 bits (160), Expect = 2e-11
Identities = 32/85 (37%), Positives = 50/85 (58%), Gaps = 3/85 (3%)
Frame = +3
Query: 38 EVRIQILFTALCHTDAYTWSGKDPEGL--FPCILGHEAAGIVESVGEGVTDVKPGDHV-I 94
+V I+IL+ +CHTD + KD G+ +P + GHE G++ VG V K GD V +
Sbjct: 141 DVTIKILYCGICHTDVH--HAKDDWGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGV 314
Query: 95 PCYQAECGECKFCKSGKTNLCGKVR 119
C A C +C++CK+ + N C K++
Sbjct: 315 GCLAASCLDCEYCKTDQENYCEKLQ 389
>BG648660 similar to SP|Q9ZRF1|MTD_ Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (64%)
Length = 721
Score = 61.2 bits (147), Expect = 6e-10
Identities = 50/189 (26%), Positives = 78/189 (40%), Gaps = 4/189 (2%)
Frame = +1
Query: 38 EVRIQILFTALCHTDAYTWSGKDPEGLFPCILGHEAAGIVESVGEGVTDVKPGDHV-IPC 96
+V +++L+ +CH+D + + +P + GHE GIV VG V K GD V + C
Sbjct: 130 DVALRVLYCGICHSDLHKAKNEWGTTNYPLVPGHEIVGIVTEVGSKVKKFKVGDRVGVGC 309
Query: 97 YQAECGECKFCKSGKTNLCGKVRAATGVGVMMNDRKPRFSIKGKPIYHFMGTST---FSQ 153
C C+ C N C ++ G ++S GTST FS
Sbjct: 310 LVDSCHSCQNCVDNLENYCPQLTLTDG---------SKYS---------DGTSTHGGFSD 435
Query: 154 YTVVHDVSVAKIHPDAALDKVCLLGCGVPTGLGAVWNTAKVEPGSIVAIFGLGTVGLAVA 213
V + V +I LD L C T + + +P + + GLG +G
Sbjct: 436 SMVSDEHFVFRIPDQLPLDAAAPLLCAGITVYSPLRHFGLDKPDMNIGVVGLGGLGHMAV 615
Query: 214 EGAKSAGAS 222
+ AK+ GA+
Sbjct: 616 KFAKAFGAN 642
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,171,744
Number of Sequences: 36976
Number of extensions: 163691
Number of successful extensions: 777
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 753
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 758
length of query: 380
length of database: 9,014,727
effective HSP length: 98
effective length of query: 282
effective length of database: 5,391,079
effective search space: 1520284278
effective search space used: 1520284278
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146819.3


