

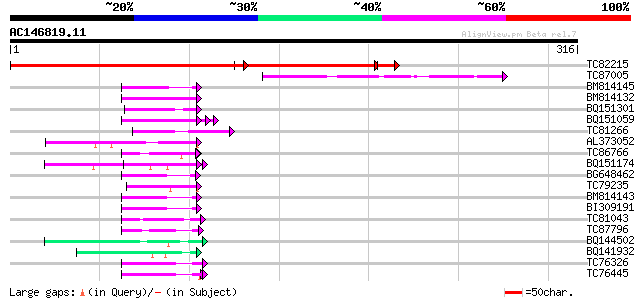
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146819.11 - phase: 0
(316 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC82215 similar to PIR|H84587|H84587 probable WD-40 repeat prote... 289 e-120
TC87005 similar to GP|9294061|dbj|BAB02018.1 WD40-repeat protein... 45 3e-05
BM814145 homologue to GP|21322711|em pherophorin-dz1 protein {Vo... 45 4e-05
BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein ... 45 4e-05
BQ151301 similar to PIR|S34666|S346 glycine-rich protein - commo... 45 5e-05
BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melano... 45 5e-05
TC81266 weakly similar to GP|16415994|emb|CAB88653. hypothetical... 44 1e-04
AL373052 similar to GP|21593014|gb| putative aldolase {Arabidops... 44 1e-04
TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E... 44 1e-04
BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xy... 43 1e-04
BG648462 weakly similar to GP|14253163|em VMP4 protein {Volvox c... 43 1e-04
TC79235 weakly similar to GP|7297741|gb|AAF52992.1| CG17108 gene... 43 2e-04
BM814143 GP|21322711|e pherophorin-dz1 protein {Volvox carteri f... 42 4e-04
BI309191 42 4e-04
TC81043 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproli... 41 5e-04
TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear prot... 41 5e-04
BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 41 5e-04
BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich gl... 41 7e-04
TC76326 homologue to PIR|C29356|C29356 hydroxyproline-rich glyco... 41 7e-04
TC76445 similar to PIR|T07623|T07623 extensin homolog HRGP2 - so... 41 7e-04
>TC82215 similar to PIR|H84587|H84587 probable WD-40 repeat protein
[imported] - Arabidopsis thaliana, partial (14%)
Length = 670
Score = 289 bits (740), Expect(3) = e-120
Identities = 133/133 (100%), Positives = 133/133 (100%)
Frame = +1
Query: 1 MEDSGEIYDGVRAEFPLSFGKQSKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHS 60
MEDSGEIYDGVRAEFPLSFGKQSKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHS
Sbjct: 16 MEDSGEIYDGVRAEFPLSFGKQSKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHS 195
Query: 61 IRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFNDSEDEDND 120
IRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFNDSEDEDND
Sbjct: 196 IRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFNDSEDEDND 375
Query: 121 SDQDELGNRFRIP 133
SDQDELGNRFRIP
Sbjct: 376 SDQDELGNRFRIP 414
Score = 154 bits (388), Expect(3) = e-120
Identities = 76/81 (93%), Positives = 76/81 (93%)
Frame = +2
Query: 126 LGNRFRIPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQ 185
L F PLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQ
Sbjct: 392 LETAFVFPLSNEIVLKGHTKVVSALAVDHTGSRVLSGSYDYMVRMYDFQGMNSRLQSFRQ 571
Query: 186 IEPSEGHQIRNLSWSPTADRF 206
IEPSEGHQIRNLSWSPTADRF
Sbjct: 572 IEPSEGHQIRNLSWSPTADRF 634
Score = 27.7 bits (60), Expect(3) = e-120
Identities = 12/12 (100%), Positives = 12/12 (100%)
Frame = +3
Query: 206 FLCVTGSAQAKI 217
FLCVTGSAQAKI
Sbjct: 633 FLCVTGSAQAKI 668
>TC87005 similar to GP|9294061|dbj|BAB02018.1 WD40-repeat protein
{Arabidopsis thaliana}, partial (74%)
Length = 1480
Score = 45.4 bits (106), Expect = 3e-05
Identities = 38/137 (27%), Positives = 64/137 (45%), Gaps = 1/137 (0%)
Frame = +3
Query: 142 GHTKVVSALAVDHTGS-RVLSGSYDYMVRMYDFQGMNSRLQSFRQIEPSEGHQIRNLSWS 200
GH++ V + A T R+++ D++V Y+ F+Q + + + +S
Sbjct: 465 GHSRRVLSCAYKPTRPFRIVTCGEDFLVNFYEGPPFR-----FKQSHRDHSNFVNCVRYS 629
Query: 201 PTADRFLCVTGSAQAKIYDRDGLTLGEFVKGDMYIRDLKNTKGHITGLTCGEWHPKTKET 260
P +F VTGS+ K DG T GE I +L + GH + W P K+
Sbjct: 630 PDGSKF--VTGSSDKKGIIFDGKT-GE------KIGELSSEGGHTGSIYAVSWSPDGKQ- 779
Query: 261 ILTSSEDGSLRIWDVND 277
+LT S D S ++WD+++
Sbjct: 780 VLTVSADKSAKVWDISE 830
>BM814145 homologue to GP|21322711|em pherophorin-dz1 protein {Volvox carteri
f. nagariensis}, partial (3%)
Length = 111
Score = 45.1 bits (105), Expect = 4e-05
Identities = 21/45 (46%), Positives = 21/45 (46%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPPH PPPPPP
Sbjct: 100 PPPPPPPPPPPPPPPPPPPPPPPPPPH------------PPPPPP 2
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 104 PPPPPPPPPPPPPPPPPPPPPPPPPP-------------PPPPPP 9
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 110 PPPPPPPPPPPPPPPPPPPPPPPPPP-------------PPPPPP 15
Score = 40.8 bits (94), Expect = 7e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 111 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPT--------PPPPPP 1
>BM814132 weakly similar to OMNI|MT3615.1 PE_PGRS family protein
{Mycobacterium tuberculosis CDC1551}, partial (7%)
Length = 164
Score = 45.1 bits (105), Expect = 4e-05
Identities = 21/45 (46%), Positives = 21/45 (46%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP A PPPPPP
Sbjct: 151 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPAPPPPPPPPPPPPPPP 17
Score = 44.7 bits (104), Expect = 5e-05
Identities = 20/45 (44%), Positives = 21/45 (46%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP +P PPPPPP PPPPPP
Sbjct: 143 PPPPPPPPPPPPPPPAPPPPPPPPPPRPPPPPPPPPPPPPPPPPP 9
Score = 44.3 bits (103), Expect = 6e-05
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 149 PPPPPPPPPPPPPPPPPAPPPPPPPPPPRPPPPPPPPPPPPPPPP 15
Score = 43.5 bits (101), Expect = 1e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 155 PPPPPPPPPPPPPPPPPPPAPPPPPPPPPPRPPPPPPPPPPPPPP 21
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 156 PPPPPPPPPPPPPPPPPPPRPPPPPPPPPPPPPPPPPPPPPPPPP 22
Score = 42.7 bits (99), Expect = 2e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 163 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPAPPPPPPPPPPP 29
Score = 40.4 bits (93), Expect = 0.001
Identities = 20/46 (43%), Positives = 20/46 (43%)
Frame = -3
Query: 62 RPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
RPP P PP PP P PPPPPP PP PPP
Sbjct: 99 RPPPPPPPPPPPPPPPPPPPPPPPPPP-------------PPAPPP 1
>BQ151301 similar to PIR|S34666|S346 glycine-rich protein - common tobacco,
partial (22%)
Length = 530
Score = 44.7 bits (104), Expect = 5e-05
Identities = 21/43 (48%), Positives = 23/43 (52%)
Frame = +2
Query: 65 KNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
K P P PP + PPPPP HEA G +I PPPPPP
Sbjct: 65 KRPKPTPPPQKQKKKKPHHPPPPPPHEA----GAVIMPPPPPP 181
Score = 36.2 bits (82), Expect = 0.017
Identities = 18/46 (39%), Positives = 24/46 (52%)
Frame = +2
Query: 62 RPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
+ P +P PP PP + + ++ PPPPPP PPPPPP
Sbjct: 107 KKPHHP-PPPPPHEAGAVIMPPPPPPP-------PPPTWHPPPPPP 220
Score = 33.9 bits (76), Expect = 0.084
Identities = 22/64 (34%), Positives = 24/64 (37%), Gaps = 19/64 (29%)
Frame = +3
Query: 63 PPKNPN-------PPSPPLQDDSPLVGP------------PPPPPHHEADDDDGEMIGPP 103
PPKN PP PP++ L P PPPPPHH PP
Sbjct: 87 PPKNKKKKNHTTPPPPPPMRRGR*LCPPRPPPPPLPHGTHPPPPPHH-----------PP 233
Query: 104 PPPP 107
PPP
Sbjct: 234 TPPP 245
Score = 32.3 bits (72), Expect = 0.25
Identities = 17/46 (36%), Positives = 18/46 (38%)
Frame = +1
Query: 65 KNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSN 110
K P PP PPPPPP G P PPPP S+
Sbjct: 67 KTQTHPPPPKTKKKKTTPPPPPPP---P*GGGGNYAPPAPPPPPSH 195
Score = 30.8 bits (68), Expect = 0.71
Identities = 15/41 (36%), Positives = 20/41 (48%)
Frame = +3
Query: 67 PNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P PP+P + PPPPP ++ + PPPPPP
Sbjct: 27 PPPPAPK*GEGGGKDPNPPPPPKNKKKKNHTT---PPPPPP 140
Score = 30.4 bits (67), Expect = 0.93
Identities = 24/80 (30%), Positives = 25/80 (31%), Gaps = 25/80 (31%)
Frame = +2
Query: 53 PSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHE--------------------- 91
P E I PP P PP PP P PPP PP H
Sbjct: 134 PPHEAGAVIMPPPPP-PPPPPTWHPPP---PPPTPPTHPPPTLFFLFFSHPFPGKKGGGG 301
Query: 92 ----ADDDDGEMIGPPPPPP 107
+ G PPPPPP
Sbjct: 302 GGEGKKERKGNKRPPPPPPP 361
Score = 28.9 bits (63), Expect = 2.7
Identities = 11/23 (47%), Positives = 14/23 (60%)
Frame = +3
Query: 84 PPPPPHHEADDDDGEMIGPPPPP 106
PPPPP + + G+ PPPPP
Sbjct: 24 PPPPPAPK*GEGGGKDPNPPPPP 92
Score = 28.9 bits (63), Expect = 2.7
Identities = 16/46 (34%), Positives = 20/46 (42%)
Frame = +3
Query: 62 RPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
+ P P PP + + PPPPPP G + PP PPP
Sbjct: 66 KDPNPPPPPKNKKKKNH--TTPPPPPPMRR-----GR*LCPPRPPP 182
Score = 27.7 bits (60), Expect = 6.0
Identities = 8/9 (88%), Positives = 9/9 (99%)
Frame = +2
Query: 83 PPPPPPHHE 91
PPPPPPHH+
Sbjct: 344 PPPPPPHHD 370
>BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melanogaster},
partial (26%)
Length = 308
Score = 44.7 bits (104), Expect = 5e-05
Identities = 21/50 (42%), Positives = 22/50 (44%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFN 112
PP P PP PP P PPPPPP PPPPPP F+
Sbjct: 174 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPGFS 25
Score = 43.9 bits (102), Expect = 8e-05
Identities = 22/54 (40%), Positives = 22/54 (40%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFNDSED 116
PP P PP PP P PPPPPP PPPPPP F D
Sbjct: 172 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPGVFPGKGD 11
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 302 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 168
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 266 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 132
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 253 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 119
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 246 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 112
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 236 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 102
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 238 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 104
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 228 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 94
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 218 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 84
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 204 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 70
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 178 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 44
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 296 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 162
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 304 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 170
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 306 PPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPPP 172
>TC81266 weakly similar to GP|16415994|emb|CAB88653. hypothetical protein
{Neurospora crassa}, partial (2%)
Length = 644
Score = 43.5 bits (101), Expect = 1e-04
Identities = 21/57 (36%), Positives = 27/57 (46%)
Frame = +3
Query: 69 PPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDSDQDE 125
PP PP + P PPPPPP + PPPPPPGS + S + + Q +
Sbjct: 471 PPPPPPSNYYPYQPPPPPPPPDNS-------YQPPPPPPGSMYYPSNNNQYNHQQQQ 620
Score = 32.3 bits (72), Expect = 0.25
Identities = 18/67 (26%), Positives = 23/67 (33%), Gaps = 25/67 (37%)
Frame = +3
Query: 70 PSPPLQDDSPLVGPPPPP-------------------------PHHEADDDDGEMIGPPP 104
P PP +P+ PPPPP HH+ + PP
Sbjct: 297 PFPPPASQNPIAPPPPPPQQRPSNWGGYGYSGVVEIPPQNSNFQHHQHQVQHHHVPHAPP 476
Query: 105 PPPGSNF 111
PPP SN+
Sbjct: 477 PPPPSNY 497
Score = 32.0 bits (71), Expect = 0.32
Identities = 21/81 (25%), Positives = 31/81 (37%), Gaps = 7/81 (8%)
Frame = +3
Query: 33 HKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPP---- 88
H+ VP + P ++ + +PP P PP Q PPPPPP
Sbjct: 438 HQVQHHHVPHAPPPPPPSNY------YPYQPPPPPPPPDNSYQ-------PPPPPPGSMY 578
Query: 89 ---HHEADDDDGEMIGPPPPP 106
++ + + PPPPP
Sbjct: 579 YPSNNNQYNHQQQQQQPPPPP 641
>AL373052 similar to GP|21593014|gb| putative aldolase {Arabidopsis
thaliana}, partial (19%)
Length = 459
Score = 43.5 bits (101), Expect = 1e-04
Identities = 34/106 (32%), Positives = 46/106 (43%), Gaps = 19/106 (17%)
Frame = +3
Query: 21 KQSKSQTPLETIHKATRRSVPTSNPK------TTSDDFPSK-----EC-LHSIRPPKNPN 68
+ K TP H + S+P+ NP +TS+ FPS +C L IR NP+
Sbjct: 3 QNQKWLTPPSLSHHQNQPSLPSLNPTPLPLPISTSNSFPSLPPLTWQCPLERIRHFSNPS 182
Query: 69 PPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPP-------PPPP 107
P +PP Q PP PPPH A + ++ P P PP
Sbjct: 183 PTTPPSQS------PPQPPPHPTATSNPVSVLMKPSTASSSSPSPP 302
>TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E8.80 -
Arabidopsis thaliana, partial (18%)
Length = 1260
Score = 43.5 bits (101), Expect = 1e-04
Identities = 18/44 (40%), Positives = 24/44 (53%)
Frame = +2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPP 106
PP +P PP PP +P+ PPPP P++ + PPPPP
Sbjct: 41 PPNSPPPPPPP----APVFSPPPPVPYYYSSPPPPPAHSPPPPP 160
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/53 (37%), Positives = 25/53 (46%), Gaps = 8/53 (15%)
Frame = +2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADD--------DDGEMIGPPPPPP 107
PP P+ P+PP+ P + PPPPPP H + PPPPPP
Sbjct: 167 PPPPPHSPTPPVY---PYLSPPPPPPVHSPPPPVYSPPPPSPPPCVEPPPPPP 316
Score = 39.3 bits (90), Expect = 0.002
Identities = 20/49 (40%), Positives = 22/49 (44%), Gaps = 4/49 (8%)
Frame = +2
Query: 63 PPKNP----NPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P +PP PP P PPPPPH + PPPPPP
Sbjct: 92 PPPVPYYYSSPPPPPAHSPPPPPXSPPPPPHSPTPPVY-PYLSPPPPPP 235
Score = 38.9 bits (89), Expect = 0.003
Identities = 19/48 (39%), Positives = 23/48 (47%), Gaps = 3/48 (6%)
Frame = +2
Query: 63 PPKNPNPPSPPLQDDSPL---VGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP +PP PP P+ + PPPPPP H + PPPP P
Sbjct: 149 PPPPXSPPPPPHSPTPPVYPYLSPPPPPPVHSPPP---PVYSPPPPSP 283
Score = 37.0 bits (84), Expect = 0.010
Identities = 21/71 (29%), Positives = 28/71 (38%), Gaps = 4/71 (5%)
Frame = +2
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPL----QDDSPLVGPPPPPPHHEADDDD 96
P P S P + H PP +P+PP P+ P+ PPP P +
Sbjct: 320 PCVEPPPPSSPAPHQTPYH---PPPSPSPPPSPVYAYPSPPPPVYTSPPPSPVYAYPSPP 490
Query: 97 GEMIGPPPPPP 107
+ PPPPP
Sbjct: 491 PPVYSSPPPPP 523
Score = 35.8 bits (81), Expect = 0.022
Identities = 24/62 (38%), Positives = 27/62 (42%), Gaps = 5/62 (8%)
Frame = +2
Query: 53 PSKECLHSIRPPK-NPNPPSPPLQDDSPLVGPPP----PPPHHEADDDDGEMIGPPPPPP 107
P +HS PP +P PPSPP + P PPP PPP A P P PP
Sbjct: 221 PPPPPVHSPPPPVYSPPPPSPPPCVEPPPPPPPPCVEPPPPSSPAPHQTPYHPPPSPSPP 400
Query: 108 GS 109
S
Sbjct: 401 PS 406
Score = 32.7 bits (73), Expect = 0.19
Identities = 26/77 (33%), Positives = 27/77 (34%), Gaps = 8/77 (10%)
Frame = +2
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPP-----SPPLQDDSPLVGPPPPPPHHEADDD 95
P S P P S PP + P SPP P V PPPPPP
Sbjct: 158 PXSPPPPPHSPTPPVYPYLSPPPPPPVHSPPPPVYSPPPPSPPPCVEPPPPPP------- 316
Query: 96 DGEMIGPP---PPPPGS 109
PP PPPP S
Sbjct: 317 ------PPCVEPPPPSS 349
Score = 32.7 bits (73), Expect = 0.19
Identities = 20/49 (40%), Positives = 23/49 (46%), Gaps = 4/49 (8%)
Frame = +2
Query: 63 PPKNP--NPPSPPLQDDSPLVGPPPPPPHHEADDDD--GEMIGPPPPPP 107
PP +P PSPP P+ PPPPP +E G PPPPP
Sbjct: 452 PPPSPVYAYPSPP----PPVYSSPPPPPVYEGPIPPVFGISYASPPPPP 586
Score = 30.8 bits (68), Expect = 0.71
Identities = 16/40 (40%), Positives = 18/40 (45%)
Frame = +2
Query: 68 NPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
+PP PP P PPPPPP + PPPP P
Sbjct: 26 SPPPPP-----PNSPPPPPPP--------APVFSPPPPVP 106
Score = 30.4 bits (67), Expect = 0.93
Identities = 17/43 (39%), Positives = 20/43 (45%)
Frame = +2
Query: 70 PSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFN 112
P+PP SP PPPPP+ PPPPPP F+
Sbjct: 2 PTPPYCVRSP----PPPPPNSP----------PPPPPPAPVFS 88
Score = 28.1 bits (61), Expect = 4.6
Identities = 19/64 (29%), Positives = 22/64 (33%), Gaps = 19/64 (29%)
Frame = +2
Query: 63 PPKNPN-----PPSPPLQDDSPLVGPP--------------PPPPHHEADDDDGEMIGPP 103
PP P PPS P +P PP PPPP + + P
Sbjct: 305 PPPPPPCVEPPPPSSPAPHQTPYHPPPSPSPPPSPVYAYPSPPPPVYTSPPPSPVYAYPS 484
Query: 104 PPPP 107
PPPP
Sbjct: 485 PPPP 496
>BQ151174 similar to GP|11036868|gb| PxORF73 peptide {Plutella xylostella
granulovirus}, partial (42%)
Length = 909
Score = 43.1 bits (100), Expect = 1e-04
Identities = 22/48 (45%), Positives = 22/48 (45%), Gaps = 4/48 (8%)
Frame = +1
Query: 64 PKNPNPPSPPLQDDSPLVGPPPP----PPHHEADDDDGEMIGPPPPPP 107
PKN PP PP P PPP PPHHE E G PPPP
Sbjct: 205 PKNNTPPPPPHTPPDPTPPQPPPQPPKPPHHEKRPRTPEPPGGRPPPP 348
Score = 42.4 bits (98), Expect = 2e-04
Identities = 31/98 (31%), Positives = 40/98 (40%), Gaps = 7/98 (7%)
Frame = +2
Query: 20 GKQSKSQTPLETIHKATRRSVPTSNP--KTTSDDFPSKECLHSIRPPKNPNPPSPPLQDD 77
G+ ++ P KA P +P K + P KE + PP+ PP P +
Sbjct: 392 GQGGRAPAPPPPPPKAREGRTPKPDPGSKENHNRTPGKEGPATHGPPEKKTPPPTPKRRG 571
Query: 78 -----SPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSN 110
+P GPPPP P G GPPPPPP N
Sbjct: 572 KHTKRAPGGGPPPPHPGRGG----GGPGGPPPPPPKKN 673
Score = 40.4 bits (93), Expect = 0.001
Identities = 26/86 (30%), Positives = 32/86 (36%)
Frame = +1
Query: 22 QSKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLV 81
Q+++ P E K R NP P PP PP+ P +P
Sbjct: 58 QARNPQPEEATKKQKRAPEKKKNPPRGQPPPPP--------PPPPQAPPTKPPTRQTPKN 213
Query: 82 GPPPPPPHHEADDDDGEMIGPPPPPP 107
PPPPPH D + PPP PP
Sbjct: 214 NTPPPPPHTPPDPTPPQ---PPPQPP 282
Score = 39.3 bits (90), Expect = 0.002
Identities = 32/123 (26%), Positives = 40/123 (32%), Gaps = 22/123 (17%)
Frame = +2
Query: 24 KSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECL--HSIRPPKNPNPPSPPLQDDSPLV 81
K +TP E H R P D P K H PP P+PP+PP +
Sbjct: 116 KRKTPPEASHPPPRPRPPRPPQPNHQHDKPQKTTHPHHPHTPPPTPHPPNPPPNHPNHPT 295
Query: 82 -----GP---------------PPPPPHHEADDDDGEMIGPPPPPPGSNFNDSEDEDNDS 121
GP P PP G PPPPPP + + D S
Sbjct: 296 TRSAHGPRNHPGDAPHRRRRPNKPGPPPRRGGGQGGRAPAPPPPPPKAREGRTPKPDPGS 475
Query: 122 DQD 124
++
Sbjct: 476 KEN 484
Score = 34.3 bits (77), Expect = 0.065
Identities = 26/103 (25%), Positives = 38/103 (36%), Gaps = 8/103 (7%)
Frame = +1
Query: 37 RRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDD 96
+ + P P T D P + +PPK P+ P + P PPPP +A
Sbjct: 208 KNNTPPPPPHTPPDPTPPQP---PPQPPKPPHHEKRPRTPEPPGGRPPPPTTPKQARPTT 378
Query: 97 GEMIG--------PPPPPPGSNFNDSEDEDNDSDQDELGNRFR 131
E G PP PP G+ D++ + + R R
Sbjct: 379 QERGGARGARPRPPPAPPQGTGGEDTKTRPGKQREPQQNPRER 507
Score = 34.3 bits (77), Expect = 0.065
Identities = 25/91 (27%), Positives = 34/91 (36%), Gaps = 14/91 (15%)
Frame = +3
Query: 28 PLETIHKATRRSV---------PTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDS 78
P HK T++S P + P + P + ++ P K + P+ P
Sbjct: 72 PTGRSHKKTKKSPRKKEKPPPRPATPPPAPAPPGPPNQTTNTTNPKKQ-HTPTTPTHPPR 248
Query: 79 PLVGPPPPP-----PHHEADDDDGEMIGPPP 104
P P PPP P EA D G G PP
Sbjct: 249 PHTPPTPPPTTQTTPPREAPTDPGTTRGTPP 341
Score = 33.1 bits (74), Expect = 0.14
Identities = 18/56 (32%), Positives = 24/56 (42%), Gaps = 11/56 (19%)
Frame = +1
Query: 63 PPKNPNPPSP---PLQDDSPLVGPPPP--------PPHHEADDDDGEMIGPPPPPP 107
PP P PP+P P + D+ P P P + + G+ PPPPPP
Sbjct: 1 PPTTPQPPAPKRPPKEKDTQARNPQPEEATKKQKRAPEKKKNPPRGQPPPPPPPPP 168
Score = 32.7 bits (73), Expect = 0.19
Identities = 16/48 (33%), Positives = 21/48 (43%)
Frame = +2
Query: 46 KTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEAD 93
K T P + H+ R P PP P + GPPPPPP ++
Sbjct: 536 KKTPPPTPKRRGKHTKRAPGGGPPPPHPGRGGGGPGGPPPPPPKKNSE 679
Score = 30.8 bits (68), Expect = 0.71
Identities = 28/108 (25%), Positives = 34/108 (30%), Gaps = 10/108 (9%)
Frame = +3
Query: 23 SKSQTPLETIHKATRRSVPTSNPKTT-SDDFPSKECLHSIRPPKNPNPPSPPLQ------ 75
S TP H + + S+ KT S K PP P PP PP Q
Sbjct: 24 SPQATPQRKGHPSQKPPTGRSHKKTKKSPRKKEKPPPRPATPPPAPAPPGPPNQTTNTTN 203
Query: 76 ---DDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSNFNDSEDEDND 120
+P PP PH PP PPP + + D
Sbjct: 204 PKKQHTPTTPTHPPRPH-----------TPPTPPPTTQTTPPREAPTD 314
Score = 29.3 bits (64), Expect = 2.1
Identities = 22/77 (28%), Positives = 28/77 (35%), Gaps = 2/77 (2%)
Frame = +3
Query: 42 TSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGP--PPPPPHHEADDDDGEM 99
T K T+ + K P K +PP + + P PPPH A G
Sbjct: 465 TREAKRTTTEPQGKRDRPRTDPRKKKHPPPHQKEGGNTQKEPRGEDPPPHTPAGGGGGPG 644
Query: 100 IGPPPPPPGSNFNDSED 116
GP PPPP E+
Sbjct: 645 -GPRPPPPKKTARGGEE 692
>BG648462 weakly similar to GP|14253163|em VMP4 protein {Volvox carteri f.
nagariensis}, partial (4%)
Length = 678
Score = 43.1 bits (100), Expect = 1e-04
Identities = 21/45 (46%), Positives = 25/45 (54%), Gaps = 1/45 (2%)
Frame = +1
Query: 63 PPKNPNPPS-PPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPP 106
PP +P+PPS PPL + P + PPPPPP G PPPP
Sbjct: 463 PPPHPHPPSQPPLPETQPPLVPPPPPP------------GSPPPP 561
Score = 35.8 bits (81), Expect = 0.022
Identities = 19/48 (39%), Positives = 22/48 (45%), Gaps = 1/48 (2%)
Frame = +1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPP-PPHHEADDDDGEMIGPPPPPPGS 109
PP PP L P + PP P PP + + PPPPPPGS
Sbjct: 406 PPSIVPPPLSSLPPPQPEMPPPHPHPPSQPPLPETQPPLVPPPPPPGS 549
Score = 27.7 bits (60), Expect = 6.0
Identities = 19/51 (37%), Positives = 22/51 (42%)
Frame = +1
Query: 53 PSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPP 103
PS+ L +PP P PP P SP PPP PP + E G P
Sbjct: 484 PSQPPLPETQPPLVPPPPPP----GSP---PPPSPPLPVQEPVSVECSGQP 615
>TC79235 weakly similar to GP|7297741|gb|AAF52992.1| CG17108 gene product
{Drosophila melanogaster}, partial (21%)
Length = 648
Score = 42.7 bits (99), Expect = 2e-04
Identities = 22/51 (43%), Positives = 24/51 (46%), Gaps = 9/51 (17%)
Frame = -2
Query: 66 NPNPPSPPLQDDSPLVGPPPPPP-----HHEADDDDGEMIGPP----PPPP 107
NP PP PP+ P +GP PPPP HH D PP PPPP
Sbjct: 512 NPPPPGPPIMGRGPPLGPGPPPPPMPHSHHPPPPHDPFAPPPPVFHHPPPP 360
Score = 36.6 bits (83), Expect = 0.013
Identities = 25/69 (36%), Positives = 28/69 (40%), Gaps = 5/69 (7%)
Frame = -2
Query: 44 NPKTTSDDF-PSKECLHSIRPPKNPNPPSPPL-QDDSPL---VGPPPPPPHHEADDDDGE 98
+P D F P H PP NP P P + +PL PPPP P H
Sbjct: 425 HPPPPHDPFAPPPPVFHHPPPPPNPYAPPPVVHHHHTPLHAHPAPPPPQPGHP------- 267
Query: 99 MIGPPPPPP 107
GPPPP P
Sbjct: 266 --GPPPPRP 246
Score = 35.4 bits (80), Expect = 0.029
Identities = 17/44 (38%), Positives = 18/44 (40%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPP 106
PP P PP PP+ PPPPH PPPPP
Sbjct: 473 PPLGPGPPPPPMPHSH-----HPPPPHDPFAPPPPVFHHPPPPP 357
Score = 33.1 bits (74), Expect = 0.14
Identities = 20/58 (34%), Positives = 22/58 (37%), Gaps = 8/58 (13%)
Frame = -2
Query: 59 HSIRPPKNPNPPSPPLQDDSPLVGPPPPPPH--------HEADDDDGEMIGPPPPPPG 108
HS PP PP P P+ PPPPP+ H PPPP PG
Sbjct: 434 HSHHPP----PPHDPFAPPPPVFHHPPPPPNPYAPPPVVHHHHTPLHAHPAPPPPQPG 273
>BM814143 GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (4%)
Length = 132
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 131 PPPPPPPPPPPPPPPPPPPPPPPPPP-------------PPPPPP 36
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 124 PPPPPPPPPPPPPPPPPPPPPPPPPP-------------PPPPPP 29
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 129 PPPPPPPPPPPPPPPPPPPPPPPPPP-------------PPPPPP 34
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 109 PPPPPPPPPPPPPPPPPPPPPPPPPP-------------PPPPPP 14
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 106 PPPPPPPPPPPPPPPPPPPPPPPPPP-------------PPPPPP 11
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 104 PPPPPPPPPPPPPPPPPPPPPPPPPP-------------PPPPPP 9
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 98 PPPPPPPPPPPPPPPPPPPPPPPPPP-------------PPPPPP 3
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 118 PPPPPPPPPPPPPPPPPPPPPPPPPP-------------PPPPPP 23
Score = 36.6 bits (83), Expect = 0.013
Identities = 19/45 (42%), Positives = 19/45 (42%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP PPPPPP PPPPPP
Sbjct: 69 PPPPPPPPPPP---------PPPPPP-------------PPPPPP 1
>BI309191
Length = 109
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -3
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 104 PPPPPPPPPPPPPPPPPPPPPPPPPP-------------PPPPPP 9
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 99 PPPPPPPPPPPPPPPPPPPPPPPPPP-------------PPPPPP 4
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 97 PPPPPPPPPPPPPPPPPPPPPPPPPP-------------PPPPPP 2
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP PPPPPP
Sbjct: 109 PPPPPPPPPPPPPPPPPPPPPPPPPP-------------PPPPPP 14
>TC81043 weakly similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich
glycoprotein DZ-HRGP {Volvox carteri f. nagariensis},
partial (19%)
Length = 555
Score = 41.2 bits (95), Expect = 5e-04
Identities = 22/47 (46%), Positives = 23/47 (48%)
Frame = +1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGS 109
PPK P PPSP +P PPP PP A G PP PPP S
Sbjct: 238 PPKPPAPPSPG-SPPAPAPAPPPIPPSPPAPPSPG---SPPSPPPAS 366
Score = 32.0 bits (71), Expect = 0.32
Identities = 26/70 (37%), Positives = 29/70 (41%), Gaps = 1/70 (1%)
Frame = +1
Query: 41 PTSNPKTTSDDFP-SKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEM 99
P S PK + P S PP P+PP+PP SP PP PPP A
Sbjct: 229 PGSPPKPPAPPSPGSPPAPAPAPPPIPPSPPAPP----SP-GSPPSPPPASPAPP----- 378
Query: 100 IGPPPPPPGS 109
P PP GS
Sbjct: 379 --KPTPPKGS 402
Score = 27.3 bits (59), Expect = 7.9
Identities = 16/49 (32%), Positives = 19/49 (38%)
Frame = +1
Query: 40 VPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPP 88
+P S P S P PP +P PP P SPL+ P P
Sbjct: 304 IPPSPPAPPSPGSPPSP------PPASPAPPKPTPPKGSPLLPMPEKTP 432
>TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear protein.
[strain B95-8 Human herpesvirus 4] {Epstein-barr
virus}, partial (23%)
Length = 431
Score = 41.2 bits (95), Expect = 5e-04
Identities = 21/46 (45%), Positives = 23/46 (49%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPG 108
PP +P P PP +P PPPPPP E PPPPPPG
Sbjct: 236 PPPSPPPIPPPY---APPCAPPPPPPPPEPP--------PPPPPPG 132
Score = 40.4 bits (93), Expect = 0.001
Identities = 19/45 (42%), Positives = 19/45 (42%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPP PP PPPPPP
Sbjct: 176 PPPPPEPPPPPPPPGDPPPYPPPVPPPLPYPTPCAPPAPPPPPPP 42
Score = 35.4 bits (80), Expect = 0.029
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P PP PP P PPPPPP D PP PPP
Sbjct: 197 PPCAPPPPPPP-----PEPPPPPPPP------GDPPPYPPPVPPP 96
Score = 35.4 bits (80), Expect = 0.029
Identities = 18/45 (40%), Positives = 20/45 (44%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP P P PPL +P P PPPP + PPPP P
Sbjct: 128 PPPYPPPVPPPLPYPTPCAPPAPPPPPPP--------LPPPPPYP 18
Score = 32.7 bits (73), Expect = 0.19
Identities = 19/46 (41%), Positives = 20/46 (43%), Gaps = 1/46 (2%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGP-PPPPPHHEADDDDGEMIGPPPPPP 107
PP P P +PP P P PPPPP D PPP PP
Sbjct: 224 PPPIPPPYAPPCAPPPPPPPPEPPPPPPPPGD--------PPPYPP 111
>BQ144502 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (39%)
Length = 1358
Score = 41.2 bits (95), Expect = 5e-04
Identities = 29/100 (29%), Positives = 36/100 (36%), Gaps = 9/100 (9%)
Frame = -3
Query: 20 GKQSKSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSP 79
G ++ S P H A RR P+ C P P PP PP P
Sbjct: 339 GSRAPSPAPPSPTHPAPRRPWTGPPPRPRPHSP*QAACGRGHPPRHRPPPPGPP---PPP 169
Query: 80 LVGPPPPP---------PHHEADDDDGEMIGPPPPPPGSN 110
+GP PPP PH + PPPPPP ++
Sbjct: 168 PLGPRPPPHPPRSGPPRPHPPTRTS----LSPPPPPPAAS 61
Score = 38.1 bits (87), Expect = 0.004
Identities = 19/45 (42%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Frame = -2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPH-HEADDDDGEMIGPPPPP 106
PP+ P PP PP P PPPPPPH H+ + PP P
Sbjct: 169 PPRPPPPPPPP-----PQRPPPPPPPHTHQPEPPTSPPRRQPPAP 50
Score = 37.4 bits (85), Expect = 0.008
Identities = 20/47 (42%), Positives = 21/47 (44%)
Frame = -1
Query: 61 IRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
+R P PP PP P V PPP PP E PPPPPP
Sbjct: 590 VRVATYPRPPRPP-----PPVPPPPSPPR---APPQSEAAPPPPPPP 474
Score = 35.8 bits (81), Expect = 0.022
Identities = 21/64 (32%), Positives = 29/64 (44%)
Frame = -2
Query: 24 KSQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGP 83
+S E++H + P +P+ P E LH+ RP P PP P +P GP
Sbjct: 604 RSSLSSESLHTPAHLAPPRQSPRPR----PPPEPLHNPRPRPPPRPP-PAAPARAPPSGP 440
Query: 84 PPPP 87
P PP
Sbjct: 439 PAPP 428
Score = 34.3 bits (77), Expect = 0.065
Identities = 20/48 (41%), Positives = 22/48 (45%), Gaps = 1/48 (2%)
Frame = -1
Query: 61 IRPPKNPNPPSPPLQDDSPLVGPP-PPPPHHEADDDDGEMIGPPPPPP 107
+RPP P PP P P PP P PPH A + PPPP P
Sbjct: 176 LRPPSAPAPPPTP-----PAAAPPAPTPPHAPA---*APHLPPPPPAP 57
Score = 33.9 bits (76), Expect = 0.084
Identities = 22/59 (37%), Positives = 26/59 (43%), Gaps = 6/59 (10%)
Frame = -2
Query: 54 SKECLHS---IRPPKN---PNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPP 106
S E LH+ + PP+ P PP PL + P P PPP A GPP PP
Sbjct: 592 SSESLHTPAHLAPPRQSPRPRPPPEPLHNPRPRPPPRPPP----AAPARAPPSGPPAPP 428
Score = 31.2 bits (69), Expect = 0.55
Identities = 18/55 (32%), Positives = 22/55 (39%), Gaps = 9/55 (16%)
Frame = -2
Query: 64 PKNPNPPSPPL---------QDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGS 109
P P+P S PL + SP PPP H + + PPPPP S
Sbjct: 850 PGPPDPASTPLDRARACCARRPPSPHRPAPPPQAHPARERRTASLARTPPPPPSS 686
Score = 29.6 bits (65), Expect = 1.6
Identities = 16/48 (33%), Positives = 18/48 (37%)
Frame = -1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSN 110
PP P PPSPP PPPP P + P P P +
Sbjct: 554 PPPVPPPPSPPRAPPQSEAAPPPP-PPPRGARPRAPLRPPSAPDPAGH 414
Score = 28.1 bits (61), Expect = 4.6
Identities = 19/65 (29%), Positives = 26/65 (39%)
Frame = -1
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMI 100
P++ P S P C + +P + PL P PPPPPP E ++
Sbjct: 791 PSALPAPPSATPPGAPCARAPHSIPRAHPAATPLL---PPPRPPPPPPEWERVPLAKKIG 621
Query: 101 GPPPP 105
G P P
Sbjct: 620 GQPRP 606
>BQ141932 weakly similar to GP|6523547|emb hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (30%)
Length = 1338
Score = 40.8 bits (94), Expect = 7e-04
Identities = 28/78 (35%), Positives = 31/78 (38%), Gaps = 8/78 (10%)
Frame = -1
Query: 38 RSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDS---PLVGPPP-----PPPH 89
RS + P PS C H PP PP PP S LV PPP PPP
Sbjct: 396 RSSTLAPPPHRQFHSPSALCTHCGIPPPAVPPPPPPTPPPSSPPALVSPPPRPFLFPPPP 217
Query: 90 HEADDDDGEMIGPPPPPP 107
++ PPPPPP
Sbjct: 216 NQPSPSP----SPPPPPP 175
Score = 37.7 bits (86), Expect = 0.006
Identities = 25/75 (33%), Positives = 33/75 (43%), Gaps = 5/75 (6%)
Frame = -3
Query: 41 PTSNPKTTSDDFPSKECLHSIRPPKNPNPPSP-----PLQDDSPLVGPPPPPPHHEADDD 95
P++ P + P + RPP P P PL+D SP PPPPPPH +
Sbjct: 445 PSAPPPSDLPPPPRSPTVIDPRPPAASAVPLPLRPLYPLRD-SPPRRPPPPPPHSPSILP 269
Query: 96 DGEMIGPPPPPPGSN 110
+ PP P P S+
Sbjct: 268 PSSRLSPPTPLPLSS 224
Score = 37.0 bits (84), Expect = 0.010
Identities = 23/69 (33%), Positives = 26/69 (37%)
Frame = -1
Query: 39 SVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGE 98
+VP P T P + PP P PP SP PPPPPP
Sbjct: 312 AVPPPPPPTPPPSSPPA----LVSPPPRPFLFPPPPNQPSPSPSPPPPPP---------- 175
Query: 99 MIGPPPPPP 107
+ PPPP P
Sbjct: 174 -LPPPPPQP 151
Score = 36.6 bits (83), Expect = 0.013
Identities = 26/91 (28%), Positives = 37/91 (40%), Gaps = 8/91 (8%)
Frame = -1
Query: 25 SQTPLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPLQDDSPLVGPP 84
SQ+P T+++ T + P P + P S+ PP P+PP+ P + P
Sbjct: 546 SQSPA-TLNRPTSHT-PACPPPSRPPPHPRAP---SLSPPPPPHPPTSPPRPGRPRSSTL 382
Query: 85 PPPPHHEADDDDG--------EMIGPPPPPP 107
PPPH + PPPPPP
Sbjct: 381 APPPHRQFHSPSALCTHCGIPPPAVPPPPPP 289
Score = 33.1 bits (74), Expect = 0.14
Identities = 20/51 (39%), Positives = 23/51 (44%), Gaps = 7/51 (13%)
Frame = -2
Query: 64 PKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPP-------PPPP 107
P++PNPP PP D P P P PPH + PP PPPP
Sbjct: 554 PQHPNPP-PP*TD--PPATPLPAPPHPVRPPTHARLRSPPLRPPTLRPPPP 411
Score = 30.4 bits (67), Expect = 0.93
Identities = 27/89 (30%), Positives = 35/89 (38%), Gaps = 6/89 (6%)
Frame = -3
Query: 28 PLETIHKATRRSVPTSNPKTTSDDFPSKECLHSIRPPKNPNPPSPPL----QDDSPLVGP 83
P TI ++R P S +T P + RP ++P PP PP + P P
Sbjct: 682 PRRTISGSSRP--PASGAGSTGHAIPYFVFGNRQRPTRHPPPP*PPSIPIPRHPEPTHQP 509
Query: 84 PP--PPPHHEADDDDGEMIGPPPPPPGSN 110
P PPP A PP PP S+
Sbjct: 508 HPCLPPPIPSAPPPTRAFALPPSAPPPSD 422
Score = 27.3 bits (59), Expect = 7.9
Identities = 14/27 (51%), Positives = 15/27 (54%), Gaps = 1/27 (3%)
Frame = -1
Query: 63 PPKNPN-PPSPPLQDDSPLVGPPPPPP 88
PP P+ PSPP PL PPP PP
Sbjct: 222 PPNQPSPSPSPP--PPPPLPPPPPQPP 148
>TC76326 homologue to PIR|C29356|C29356 hydroxyproline-rich glycoprotein
(clone Hyp2.13) - kidney bean (fragment), partial (32%)
Length = 777
Score = 40.8 bits (94), Expect = 7e-04
Identities = 19/48 (39%), Positives = 23/48 (47%)
Frame = +1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSN 110
PP +P+PP P P P PPPP+H PPPP P S+
Sbjct: 109 PPPSPSPPPPYYYKSPPPPSPSPPPPYHYQS--------PPPPSPISH 228
Score = 38.9 bits (89), Expect = 0.003
Identities = 19/52 (36%), Positives = 23/52 (43%), Gaps = 5/52 (9%)
Frame = +1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPP-----PPPGS 109
PP +P+PP P P P PPPP++ PPP PPP S
Sbjct: 61 PPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYHYQSPPPPS 216
Score = 38.9 bits (89), Expect = 0.003
Identities = 19/52 (36%), Positives = 23/52 (43%), Gaps = 5/52 (9%)
Frame = +1
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPP-----PPPGS 109
PP +P+PP P P P PPPP++ PPP PPP S
Sbjct: 13 PPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPS 168
Score = 32.3 bits (72), Expect = 0.25
Identities = 24/73 (32%), Positives = 28/73 (37%), Gaps = 25/73 (34%)
Frame = +1
Query: 63 PPKNPNPP------SPPLQDDSPLVGPP------------PPPPHHEADDDDGEMIGPP- 103
PP +P+PP SPP SP+ PP PPPP+H PP
Sbjct: 157 PPPSPSPPPPYHYQSPP--PPSPISHPPYYYKSPPPPSPSPPPPYHYVSPPPPVKSPPPP 330
Query: 104 ------PPPPGSN 110
PPPP N
Sbjct: 331 TYIYASPPPPHYN 369
Score = 31.2 bits (69), Expect = 0.55
Identities = 21/67 (31%), Positives = 28/67 (41%), Gaps = 7/67 (10%)
Frame = +1
Query: 53 PSKECLHSIRPPKNPNPPS-PPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPP------ 105
PS + + P P+P S PP SP P PPP + + PPPP
Sbjct: 169 PSPPPPYHYQSPPPPSPISHPPYYYKSPPPPSPSPPPPYHYVSPPPPVKSPPPPTYIYAS 348
Query: 106 PPGSNFN 112
PP ++N
Sbjct: 349 PPPPHYN 369
Score = 27.3 bits (59), Expect = 7.9
Identities = 14/41 (34%), Positives = 14/41 (34%)
Frame = +1
Query: 67 PNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
P PP P P PPPP P PPPP
Sbjct: 4 PGPPPPSPSPPPPYYYKSPPPP------------SPSPPPP 90
>TC76445 similar to PIR|T07623|T07623 extensin homolog HRGP2 - soybean
(fragment), partial (76%)
Length = 821
Score = 40.8 bits (94), Expect = 7e-04
Identities = 19/48 (39%), Positives = 23/48 (47%)
Frame = +2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPPGSN 110
PP +P+PP P P P PPPP+H PPPP P S+
Sbjct: 134 PPPSPSPPPPYYYKSPPPPSPSPPPPYHYQS--------PPPPSPISH 253
Score = 40.8 bits (94), Expect = 7e-04
Identities = 20/52 (38%), Positives = 24/52 (45%), Gaps = 5/52 (9%)
Frame = +2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPP-----PPPGS 109
PP +P+PPSP P P PPPP++ PPP PPP S
Sbjct: 38 PPPSPSPPSPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPS 193
Score = 38.9 bits (89), Expect = 0.003
Identities = 19/52 (36%), Positives = 23/52 (43%), Gaps = 5/52 (9%)
Frame = +2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPP-----PPPGS 109
PP +P+PP P P P PPPP++ PPP PPP S
Sbjct: 86 PPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYHYQSPPPPS 241
Score = 33.1 bits (74), Expect = 0.14
Identities = 19/45 (42%), Positives = 22/45 (48%)
Frame = +2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPPPP 107
PP +P+PP PP SP PPP P H + PPPP P
Sbjct: 182 PPPSPSPP-PPYHYQSP---PPPSPISHPPN----YYKSPPPPSP 292
Score = 32.3 bits (72), Expect = 0.25
Identities = 15/43 (34%), Positives = 18/43 (40%)
Frame = +2
Query: 63 PPKNPNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPP 105
PP +P P P P PPPP+H + PPPP
Sbjct: 230 PPPSPISHPPNYYKSPPPPSPSPPPPYH--------YVSPPPP 334
Score = 30.4 bits (67), Expect = 0.93
Identities = 16/48 (33%), Positives = 19/48 (39%), Gaps = 5/48 (10%)
Frame = +2
Query: 67 PNPPSPPLQDDSPLVGPPPPPPHHEADDDDGEMIGPPP-----PPPGS 109
P+PP P P P PP P++ PPP PPP S
Sbjct: 2 PSPPPPYYYKSPPPPSPSPPSPYYYKSPPPPSPSPPPPYYYKSPPPPS 145
Score = 29.6 bits (65), Expect = 1.6
Identities = 17/44 (38%), Positives = 20/44 (44%), Gaps = 1/44 (2%)
Frame = +2
Query: 63 PPKNPNPPSP-PLQDDSPLVGPPPPPPHHEADDDDGEMIGPPPP 105
PP +P+PP P P V PPPP + A PPPP
Sbjct: 278 PPPSPSPPPPYHYVSPPPPVKSPPPPAYIYA--------SPPPP 385
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,279,774
Number of Sequences: 36976
Number of extensions: 290606
Number of successful extensions: 10207
Number of sequences better than 10.0: 566
Number of HSP's better than 10.0 without gapping: 3220
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5772
length of query: 316
length of database: 9,014,727
effective HSP length: 96
effective length of query: 220
effective length of database: 5,465,031
effective search space: 1202306820
effective search space used: 1202306820
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146819.11


