

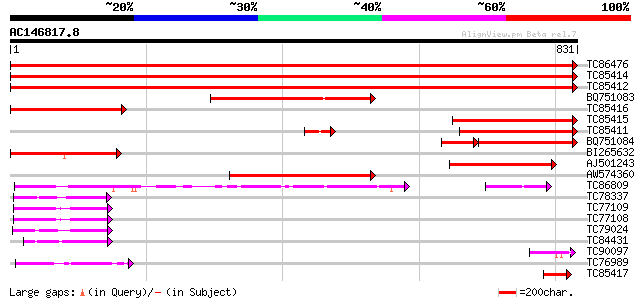
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146817.8 + phase: 0
(831 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86476 similar to GP|15450763|gb|AAK96653.1 elongation factor E... 1670 0.0
TC85414 homologue to GP|15450763|gb|AAK96653.1 elongation factor... 1483 0.0
TC85412 homologue to GP|15450763|gb|AAK96653.1 elongation factor... 1476 0.0
BQ751083 homologue to GP|13925370|gb elongation factor 2 {Neuros... 321 6e-88
TC85416 homologue to GP|15450763|gb|AAK96653.1 elongation factor... 309 3e-84
TC85415 homologue to SP|O23755|EF2_BETVU Elongation factor 2 (EF... 306 2e-83
TC85411 homologue to SP|O23755|EF2_BETVU Elongation factor 2 (EF... 288 5e-78
BQ751084 homologue to GP|13925370|gb elongation factor 2 {Neuros... 184 3e-61
BI265632 homologue to GP|22947038|gb CG2238-PA {Drosophila melan... 222 5e-58
AJ501243 weakly similar to PIR|A40411|A40 translation elongation... 206 2e-53
AW574360 similar to PIR|H86197|H86 hypothetical protein [importe... 167 1e-41
TC86809 homologue to PIR|S35701|S35701 translation elongation fa... 133 3e-31
TC78337 similar to GP|9759359|dbj|BAB10014.1 GTP-binding protein... 102 5e-22
TC77109 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein... 99 5e-21
TC77108 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein... 99 5e-21
TC79024 similar to PIR|A84716|A84716 probable GTP-binding protei... 95 1e-19
TC84431 similar to GP|10176983|dbj|BAB10215. GTP-binding membran... 87 2e-17
TC90097 homologue to PIR|H86197|H86197 hypothetical protein [imp... 59 8e-09
TC76989 homologue to SP|O24310|EFTU_PEA Elongation factor Tu ch... 57 2e-08
TC85417 similar to SP|Q39211|RP3A_ARATH DNA-directed RNA polymer... 51 2e-06
>TC86476 similar to GP|15450763|gb|AAK96653.1 elongation factor EF-2
{Arabidopsis thaliana}, complete
Length = 2974
Score = 1670 bits (4326), Expect = 0.0
Identities = 831/831 (100%), Positives = 831/831 (100%)
Frame = +2
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS
Sbjct: 281 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 460
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV
Sbjct: 461 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 640
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY
Sbjct: 641 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 820
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS
Sbjct: 821 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 1000
Query: 241 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL 300
STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL
Sbjct: 1001 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL 1180
Query: 301 SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP 360
SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP
Sbjct: 1181 SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP 1360
Query: 361 EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT 420
EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT
Sbjct: 1361 EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT 1540
Query: 421 VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVA 480
VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVA
Sbjct: 1541 VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVA 1720
Query: 481 VTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNG 540
VTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNG
Sbjct: 1721 VTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNG 1900
Query: 541 AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE 600
AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE
Sbjct: 1901 AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE 2080
Query: 601 PKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIAS 660
PKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIAS
Sbjct: 2081 PKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIAS 2260
Query: 661 KEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLV 720
KEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLV
Sbjct: 2261 KEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLV 2440
Query: 721 EIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQ 780
EIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQ
Sbjct: 2441 EIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQ 2620
Query: 781 AFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
AFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL
Sbjct: 2621 AFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 2773
>TC85414 homologue to GP|15450763|gb|AAK96653.1 elongation factor EF-2
{Arabidopsis thaliana}, complete
Length = 2892
Score = 1483 bits (3840), Expect = 0.0
Identities = 720/831 (86%), Positives = 775/831 (92%)
Frame = +3
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD KHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTR DEAERGITIKS
Sbjct: 162 MDYKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKS 341
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISLYYEM+D LK+FKGER GN+YLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV
Sbjct: 342 TGISLYYEMTDDSLKSFKGERNGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 521
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQALGERI+PVLTVNKMDRCFLEL +D EEAY T RVIE+ NV+MATY
Sbjct: 522 EGVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVDGEEAYQTFSRVIENANVIMATY 701
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
ED LLGDVQVYPEKGTV+FSAGLHGW+FTLTNFAKMYASKFGVDE KMM RLWGENFFD
Sbjct: 702 EDPLLGDVQVYPEKGTVAFSAGLHGWAFTLTNFAKMYASKFGVDETKMMERLWGENFFDP 881
Query: 241 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL 300
+TKKWT K+T + TCKRGFVQFCYEPIKQ+I CMNDQKDKLWPML KLG+ +KSEEK+L
Sbjct: 882 ATKKWTTKNTGSATCKRGFVQFCYEPIKQVINTCMNDQKDKLWPMLTKLGITMKSEEKDL 1061
Query: 301 SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP 360
GK LMKRVMQ+WLPAS+ALLEMMIFHLPSP+ AQ+YRVENLYEGPLDD YA+AIRNCDP
Sbjct: 1062MGKPLMKRVMQTWLPASTALLEMMIFHLPSPSTAQRYRVENLYEGPLDDQYATAIRNCDP 1241
Query: 361 EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT 420
EGPLMLYVSKMIPASDKGRF+AFGRVFSGKVSTG+KVRIMGPN++PGEKKDLYVKSVQRT
Sbjct: 1242EGPLMLYVSKMIPASDKGRFFAFGRVFSGKVSTGLKVRIMGPNFVPGEKKDLYVKSVQRT 1421
Query: 421 VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVA 480
VIWMGK+QETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVV VA
Sbjct: 1422VIWMGKRQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVRVA 1601
Query: 481 VTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNG 540
V CKVASDLPKLVEGLKRLAKSDPMVVCTI E+GEHI+A AGELHLEICLKDLQDDFM G
Sbjct: 1602VQCKVASDLPKLVEGLKRLAKSDPMVVCTIEESGEHIVAGAGELHLEICLKDLQDDFMGG 1781
Query: 541 AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE 600
AEI KSDP+VSFRETVLE+S TVMSKSPNKHNRLYMEARP+E+GLAEAIDDG+IGPRD+
Sbjct: 1782AEIIKSDPVVSFRETVLERSCRTVMSKSPNKHNRLYMEARPLEDGLAEAIDDGKIGPRDD 1961
Query: 601 PKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIAS 660
PKN KILS+E+GWDKDLAKK+WCFGPETTGPNM+VD CKGVQYLNEIKDSVVAGFQ AS
Sbjct: 1962PKNRSKILSEEYGWDKDLAKKIWCFGPETTGPNMVVDMCKGVQYLNEIKDSVVAGFQWAS 2141
Query: 661 KEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLV 720
KEG +++EN+R +CFEVCDVVLHTDAIHRGGGQIIPTARRVFYA+ LTAKPRLLEPVY+V
Sbjct: 2142KEGALSEENMRAICFEVCDVVLHTDAIHRGGGQIIPTARRVFYASQLTAKPRLLEPVYMV 2321
Query: 721 EIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQ 780
EIQAPE ALGGIYSVLNQKRGHVF+E+QRP TPLYN+KAYLPVIESF F+ LRA T GQ
Sbjct: 2322EIQAPEQALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVIESFGFSSQLRAATSGQ 2501
Query: 781 AFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
AFPQ VFDHWDM+ SDPLE G+ AA V +IRK+KGLKEQ+ PLSEFED+L
Sbjct: 2502AFPQCVFDHWDMMSSDPLEAGSQAATLVTDIRKRKGLKEQMTPLSEFEDKL 2654
>TC85412 homologue to GP|15450763|gb|AAK96653.1 elongation factor EF-2
{Arabidopsis thaliana}, complete
Length = 3229
Score = 1476 bits (3820), Expect = 0.0
Identities = 718/831 (86%), Positives = 772/831 (92%)
Frame = +3
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD KHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTR DEAERGITIKS
Sbjct: 204 MDYKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKS 383
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISLYYEM+D LK FKGER GN+YLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV
Sbjct: 384 TGISLYYEMTDESLKRFKGERNGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 563
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATY 180
EGVCVQTETVLRQALGERI+PVLTVNKMDRCFLEL +D EEAY T RVIE+ NV+MATY
Sbjct: 564 EGVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVDGEEAYQTFSRVIENANVIMATY 743
Query: 181 EDALLGDVQVYPEKGTVSFSAGLHGWSFTLTNFAKMYASKFGVDEEKMMNRLWGENFFDS 240
ED LLGDVQVYPEKGTV+FSAGLHGW+FTLTNFAKMYASKFGVDE KMM RLWGENFFD
Sbjct: 744 EDPLLGDVQVYPEKGTVAFSAGLHGWAFTLTNFAKMYASKFGVDESKMMERLWGENFFDP 923
Query: 241 STKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELCMNDQKDKLWPMLQKLGVNLKSEEKEL 300
+TKKWT K+T + +CKRGFVQFCYEPIKQII CMNDQKDKLWPML KLGV +KSEEK+L
Sbjct: 924 ATKKWTTKNTGSASCKRGFVQFCYEPIKQIINTCMNDQKDKLWPMLTKLGVTMKSEEKDL 1103
Query: 301 SGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDP 360
GK LMKRVMQ+WLPAS+ALLEMMIFHLPSP+ AQ+YRVENLYEGPLDD YA+AIRNCDP
Sbjct: 1104 MGKPLMKRVMQTWLPASTALLEMMIFHLPSPSTAQRYRVENLYEGPLDDQYANAIRNCDP 1283
Query: 361 EGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRT 420
EGPLMLYVSKMIPASDKGRF+AFGRVF+GKVSTG+KVRIMGPNYIPGEKKDLY KSVQRT
Sbjct: 1284 EGPLMLYVSKMIPASDKGRFFAFGRVFAGKVSTGLKVRIMGPNYIPGEKKDLYTKSVQRT 1463
Query: 421 VIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVSVA 480
VIWMGK+QETVEDVPCGNTVA+VGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVV VA
Sbjct: 1464 VIWMGKRQETVEDVPCGNTVALVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVVRVA 1643
Query: 481 VTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQDDFMNG 540
V CKVASDLPKLVEGLKRLAKSDPMVVCTI E+GEHI+A AGELHLEICLKDLQDDFM G
Sbjct: 1644 VQCKVASDLPKLVEGLKRLAKSDPMVVCTIEESGEHIVAGAGELHLEICLKDLQDDFMGG 1823
Query: 541 AEITKSDPIVSFRETVLEKSSHTVMSKSPNKHNRLYMEARPMEEGLAEAIDDGRIGPRDE 600
AEI KSDP+VSFRETVL++S TVMSKSPNKHNRLYMEARP+EEGLAEAID+G IGPRD+
Sbjct: 1824 AEIIKSDPVVSFRETVLDRSVRTVMSKSPNKHNRLYMEARPLEEGLAEAIDEGTIGPRDD 2003
Query: 601 PKNHLKILSDEFGWDKDLAKKVWCFGPETTGPNMLVDTCKGVQYLNEIKDSVVAGFQIAS 660
PKN KILS+++GWDKDLAKK+WCFGPETTGPNM+VD CKGVQYLNEIKDSVVAGFQ AS
Sbjct: 2004 PKNRSKILSEQYGWDKDLAKKIWCFGPETTGPNMVVDMCKGVQYLNEIKDSVVAGFQWAS 2183
Query: 661 KEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLV 720
KEG +++EN+RG+CFEVCDVVLHTDAIHRGGGQIIPTARRVFYA+ LTAKPRLLEPVY+V
Sbjct: 2184 KEGALSEENMRGICFEVCDVVLHTDAIHRGGGQIIPTARRVFYASQLTAKPRLLEPVYMV 2363
Query: 721 EIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQ 780
EIQAPE ALGGIYSVLNQKRGHVF+E+QRP TPLYN+KAYLPVIESF F+ LRA T GQ
Sbjct: 2364 EIQAPEQALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVIESFGFSSQLRAATSGQ 2543
Query: 781 AFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
AFPQ VFDHWD + SDPLE G+ AA V +IRK+KGLKEQ+ PLSEFED+L
Sbjct: 2544 AFPQCVFDHWDTMTSDPLEAGSQAAQLVTDIRKRKGLKEQMTPLSEFEDKL 2696
>BQ751083 homologue to GP|13925370|gb elongation factor 2 {Neurospora
crassa}, partial (28%)
Length = 721
Score = 321 bits (823), Expect = 6e-88
Identities = 155/241 (64%), Positives = 194/241 (80%)
Frame = +3
Query: 295 SEEKELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKAQKYRVENLYEGPLDDPYASA 354
+E+KE GK L+K VM+++LPA+ ALLEMMI HLPSP AQKYR E LYEGP DD A A
Sbjct: 3 TEDKEKEGKQLLKAVMRTFLPAADALLEMMIIHLPSPVTAQKYRSETLYEGPPDDEAAIA 182
Query: 355 IRNCDPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYV 414
IR+CDP+GPLMLYVSKM+P SDKGRFYAFGRVF+G V +G+KVRI GPNY+PG+K+DL++
Sbjct: 183 IRDCDPKGPLMLYVSKMVPTSDKGRFYAFGRVFAGTVRSGLKVRIQGPNYVPGKKEDLFI 362
Query: 415 KSVQRTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVS 474
K++QRTV+ MG K E ++D+P GN V +VG+DQF+ K+ TLT AH ++ MKFSVS
Sbjct: 363 KAIQRTVLMMGGKVEPIDDMPAGNIVGLVGIDQFLLKSGTLTTSD--TAHNLKVMKFSVS 536
Query: 475 PVVSVAVTCKVASDLPKLVEGLKRLAKSDPMVVCTISETGEHIIAAAGELHLEICLKDLQ 534
PVV +V K A DLPKLVEGLKRL+KSDP V+ +E+GEH++A AGELHLEICL DL+
Sbjct: 537 PVVQRSVQVKNAQDLPKLVEGLKRLSKSDPCVLTMTNESGEHVVAGAGELHLEICLNDLE 716
Query: 535 D 535
+
Sbjct: 717 N 719
>TC85416 homologue to GP|15450763|gb|AAK96653.1 elongation factor EF-2
{Arabidopsis thaliana}, partial (21%)
Length = 660
Score = 309 bits (791), Expect = 3e-84
Identities = 157/171 (91%), Positives = 161/171 (93%)
Frame = +3
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD KHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTR DEAERGITIKS
Sbjct: 147 MDYKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKS 326
Query: 61 TGISLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 120
TGISLYYEMSD LK+FKGER GN+YLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV
Sbjct: 327 TGISLYYEMSDESLKSFKGERNGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCV 506
Query: 121 EGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAYSTIQRVIE 171
EGVCVQTETVLRQALGERI+PVLTVNKMDRCFLEL +D EEAY T RVIE
Sbjct: 507 EGVCVQTETVLRQALGERIRPVLTVNKMDRCFLELQVDGEEAYQTFSRVIE 659
>TC85415 homologue to SP|O23755|EF2_BETVU Elongation factor 2 (EF-2). [Sugar
beet] {Beta vulgaris}, partial (21%)
Length = 725
Score = 306 bits (785), Expect = 2e-83
Identities = 147/183 (80%), Positives = 164/183 (89%)
Frame = +1
Query: 649 KDSVVAGFQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLT 708
KDSVVAGFQ ASKEG +++EN+R +CFEVCDVVLHTDAIHRGGGQIIPTARRVFYA+ LT
Sbjct: 1 KDSVVAGFQWASKEGALSEENMRAICFEVCDVVLHTDAIHRGGGQIIPTARRVFYASQLT 180
Query: 709 AKPRLLEPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQ 768
AKPRLLEPVY+VEIQAPE ALGGIYSVLNQKRGHVF+E+QRP TPLYN+KAYLPVIESF
Sbjct: 181 AKPRLLEPVYMVEIQAPEQALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVIESFG 360
Query: 769 FNESLRAQTGGQAFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFE 828
F+ LRA T GQAFPQ VFDHWDM+ SDPLE G+ AA V +IRK+KGLKEQ+ PLSEFE
Sbjct: 361 FSSQLRAATSGQAFPQCVFDHWDMMSSDPLEAGSQAATLVTDIRKRKGLKEQMTPLSEFE 540
Query: 829 DRL 831
D+L
Sbjct: 541 DKL 549
>TC85411 homologue to SP|O23755|EF2_BETVU Elongation factor 2 (EF-2). [Sugar
beet] {Beta vulgaris}, partial (23%)
Length = 842
Score = 288 bits (738), Expect = 5e-78
Identities = 137/172 (79%), Positives = 154/172 (88%)
Frame = +2
Query: 660 SKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYL 719
SKEG +++EN+RG+CFEVCDVVLHTDAIHRGGGQIIPTARRVFYA+ LTAKPRLLEPVY+
Sbjct: 71 SKEGAVSEENMRGICFEVCDVVLHTDAIHRGGGQIIPTARRVFYASQLTAKPRLLEPVYM 250
Query: 720 VEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVIESFQFNESLRAQTGG 779
VEIQAPE ALGGIYSVLNQKRGHVF+E+QRP TPLYN+KAYLPVIESF F+ LRA T G
Sbjct: 251 VEIQAPEQALGGIYSVLNQKRGHVFEEMQRPGTPLYNIKAYLPVIESFGFSSQLRAATSG 430
Query: 780 QAFPQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIPLSEFEDRL 831
QAFPQ VFDHWD + SDPLE G+ AA V +IRK+KGLKEQ+ PLSEFED+L
Sbjct: 431 QAFPQCVFDHWDTMTSDPLEAGSQAAQLVTDIRKRKGLKEQMTPLSEFEDKL 586
Score = 52.8 bits (125), Expect = 5e-07
Identities = 28/46 (60%), Positives = 32/46 (68%)
Frame = +2
Query: 432 EDVPCGNTVAMVGLDQFITKNATLTNEKEVDAHPIRAMKFSVSPVV 477
EDVPCGNTVA+VGLDQFIT NAT + E V +R + F V VV
Sbjct: 2 EDVPCGNTVALVGLDQFITNNAT-SKEGAVSEENMRGICFEVCDVV 136
>BQ751084 homologue to GP|13925370|gb elongation factor 2 {Neurospora
crassa}, partial (23%)
Length = 761
Score = 184 bits (468), Expect(2) = 3e-61
Identities = 89/146 (60%), Positives = 110/146 (74%), Gaps = 1/146 (0%)
Frame = -3
Query: 687 IHRGGGQIIPTARRVFYAAMLTAKPRLLEPVYLVEIQAPEHALGGIYSVLNQKRGHVFDE 746
IHRGGGQIIPTARRV YAA L A+P LLEPV+LVEIQ PE A+GG+Y VL ++RGHVF E
Sbjct: 597 IHRGGGQIIPTARRVLYAAALLAEPALLEPVFLVEIQVPEQAMGGVYGVLTRRRGHVFGE 418
Query: 747 IQRPNTPLYNVKAYLPVIESFQFNESLRAQTGGQAFPQLVFDHWDMVP-SDPLEPGTPAA 805
QRP TPL+ +KAYLPV+ESF FN LR T GQAFPQ VFDHW ++P PL+P +
Sbjct: 417 EQRPGTPLFTIKAYLPVMESFGFNGDLRQATSGQAFPQSVFDHWQVLPGGSPLDPTSKVG 238
Query: 806 ARVVEIRKKKGLKEQLIPLSEFEDRL 831
V E+RK+KG+K ++ + + D+L
Sbjct: 237 TIVQEMRKRKGIKVEVPGVDNYYDKL 160
Score = 70.1 bits (170), Expect(2) = 3e-61
Identities = 32/53 (60%), Positives = 42/53 (78%)
Frame = -2
Query: 634 MLVDTCKGVQYLNEIKDSVVAGFQIASKEGPMADENLRGVCFEVCDVVLHTDA 686
+LVD + VQYLNEIKDSVV+GFQ A++EGP+A+E +R + F + DV LH DA
Sbjct: 757 LLVDQTQAVQYLNEIKDSVVSGFQWATREGPVAEEPMRSIRFNIMDVTLHADA 599
>BI265632 homologue to GP|22947038|gb CG2238-PA {Drosophila melanogaster},
partial (21%)
Length = 593
Score = 222 bits (565), Expect = 5e-58
Identities = 117/167 (70%), Positives = 132/167 (78%), Gaps = 4/167 (2%)
Frame = +3
Query: 1 MDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKS 60
MD K NIRNMSVIAHVDHGKSTLTDSLV+ AGIIA AG+ R TDTR+DE +R ITIKS
Sbjct: 93 MDKKRNIRNMSVIAHVDHGKSTLTDSLVSKAGIIAGARAGETRFTDTRKDEQDRCITIKS 272
Query: 61 TGISLYYEMSDGDLKNFKG----EREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVV 116
T IS+++E+ + DL E+ +LINLIDSPGHVDFSSEVTAALR+TDGALVV
Sbjct: 273 TAISMFFELEEKDLVFITNPDQREKSEKGFLINLIDSPGHVDFSSEVTAALRVTDGALVV 452
Query: 117 VDCVEGVCVQTETVLRQALGERIKPVLTVNKMDRCFLELHLDAEEAY 163
VDCV GVCVQTETVLRQA+ ERIKP L +NKMDR LEL L+AE Y
Sbjct: 453 VDCVSGVCVQTETVLRQAIAERIKPXLFMNKMDRALLELQLEAEXLY 593
>AJ501243 weakly similar to PIR|A40411|A40 translation elongation factor
eEF-2 - Caenorhabditis elegans, partial (18%)
Length = 497
Score = 206 bits (525), Expect = 2e-53
Identities = 90/157 (57%), Positives = 121/157 (76%)
Frame = +1
Query: 645 LNEIKDSVVAGFQIASKEGPMADENLRGVCFEVCDVVLHTDAIHRGGGQIIPTARRVFYA 704
++EIKDSVV+ FQ A+KEG + E +RG+ + + +V LH DA+HRGG QIIPTARR FYA
Sbjct: 13 MSEIKDSVVSAFQWAAKEGVLCSEGMRGIRYNIVEVSLHADAVHRGGSQIIPTARRAFYA 192
Query: 705 AMLTAKPRLLEPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAYLPVI 764
A L+A+PRL+EP++L +IQ P+ A+GG+YSV+N++RG + +E+QRP TP+ N++ YLPV
Sbjct: 193 AQLSAQPRLMEPIFLADIQCPQSAVGGVYSVMNKRRGQILEELQRPGTPMLNIRCYLPVT 372
Query: 765 ESFQFNESLRAQTGGQAFPQLVFDHWDMVPSDPLEPG 801
ESF F LR T GQAFPQ VFDHW ++ DP E G
Sbjct: 373 ESFGFTADLRGSTSGQAFPQCVFDHWKVISDDPYEKG 483
>AW574360 similar to PIR|H86197|H86 hypothetical protein [imported] -
Arabidopsis thaliana, partial (22%)
Length = 670
Score = 167 bits (424), Expect = 1e-41
Identities = 83/214 (38%), Positives = 132/214 (60%), Gaps = 1/214 (0%)
Frame = +3
Query: 323 MMIFHLPSPTKAQKYRVENLYEGPLDDPYASAIRNCDPEGPLMLYVSKMIPASDKGRFYA 382
M++ H+PSP A +V+++Y GP D A+ CD GPLM+ ++K+ P SD F A
Sbjct: 3 MLVQHIPSPRDAAVKKVDHIYTGPKDSSIYKAMTQCDSSGPLMVNITKLYPKSDCSVFDA 182
Query: 383 FGRVFSGKVSTGMKVRIMGPNYIPGEKKDLYVKSVQRTVIWMGKKQETVEDVPCGNTVAM 442
FGRV+SGK+ TG VR++G Y P +++D+ VK V + ++ + + + + P G+ V +
Sbjct: 183 FGRVYSGKIQTGQTVRVLGEGYSPDDEEDMTVKEVTKLWVYQARDRMPIAEAPPGSWVLI 362
Query: 443 VGLDQFITKNATLTN-EKEVDAHPIRAMKFSVSPVVSVAVTCKVASDLPKLVEGLKRLAK 501
G+D I K ATL N + + D + R + F+ VV A S+LPK+VEGL++++K
Sbjct: 363 EGVDASIMKTATLCNVDFDEDVYIFRPLLFNTLSVVKTATEPLNPSELPKMVEGLRKISK 542
Query: 502 SDPMVVCTISETGEHIIAAAGELHLEICLKDLQD 535
S P+ V + E+GEH I GEL+L+ +KDL++
Sbjct: 543 SYPLAVTKVEESGEHTILXTGELYLDSIMKDLRE 644
>TC86809 homologue to PIR|S35701|S35701 translation elongation factor EF-G
chloroplast - soybean, partial (91%)
Length = 2674
Score = 133 bits (334), Expect = 3e-31
Identities = 155/616 (25%), Positives = 250/616 (40%), Gaps = 38/616 (6%)
Frame = +2
Query: 8 RNMSVIAHVDHGKSTLTDSLVAAAGIIAQ--EVAGDVRMTDTRQDEAERGITIKSTGISL 65
RN+ ++AH+D GK+T T+ ++ G + EV D + E ERGITI S +
Sbjct: 320 RNIGIMAHIDAGKTTTTERILFYTGRNYKIGEVHEGTATMDWMEQEQERGITITSAATTT 499
Query: 66 YYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCV 125
+++ + IN+ID+PGHVDF+ EV ALR+ DGA+ + D V GV
Sbjct: 500 FWD----------------NHRINIIDTPGHVDFTLEVERALRVLDGAICLFDSVAGVEP 631
Query: 126 QTETVLRQALGERIKPVLTVNKMDR--------------------CFLELHLDAEEAYST 165
Q+ETV RQA + + VNKMDR L+L + AE+++
Sbjct: 632 QSETVWRQADRYGVPRICFVNKMDRLGANFFRTRDMIVTNLGAKPLVLQLPIGAEDSFKG 811
Query: 166 IQRVIESVNVVMA--------TYEDA---LLGDVQVYPEKGTVSFSAGLHGWSFTLTNFA 214
+ ++ +V TYED LL Q Y +
Sbjct: 812 VIDLVRMKAIVWGGEELGAKFTYEDIPVDLLEQAQDYR---------------------S 928
Query: 215 KMYASKFGVDEEKMMNRLWGENFFDSSTKKWTNKHTSTPTCKRGFVQFCYEPIKQIIELC 274
+M + +D+E M N L G +++ KK ++G + + P+
Sbjct: 929 QMIETIVELDDEAMENYLEGVEPDEATIKK---------LIRKGSIAATFVPV------- 1060
Query: 275 MNDQKDKLWPMLQKLGVNLKSEEKELSGKALMKRVMQSWLPASSALLEMMIFHLPSPTKA 334
+ G A + +Q LL+ ++ +LPSP
Sbjct: 1061-------------------------MCGSAFKNKGVQ-------PLLDAVVDYLPSPLD- 1141
Query: 335 QKYRVENLYEGPLDDPYASAIRNCDPEGPLMLYVSKMIPASDKGRFYAFGRVFSGKVSTG 394
V + ++P A+ R + P K++ S G F RV+SGK++ G
Sbjct: 1142----VPPMKGTDPENPEATIERIAGDDEPFSGLAFKIMSDSFVGSL-TFVRVYSGKLTAG 1306
Query: 395 MKVRIMGPNYIPGEKKDLYVKSVQRTVIWMGKKQETVEDVPCGNTVAMVGLDQFITKNAT 454
V N G+K + + R + +E V+ G+ VA+ GL IT
Sbjct: 1307SYVL----NSNKGKK-----ERIGRLLEMHANSREDVKVALTGDIVALAGLKDTITGETL 1459
Query: 455 LTNEKEVDAHPIRAMKFSVSPVVSVAVTCKVASDLPKLVEGLKRLAKSDPMV-VCTISET 513
E V + M F PV+ +A+ K +D+ K+ GL +LA+ DP E
Sbjct: 1460CDPESPV---VLERMDFP-DPVIKIAIEPKTKADIDKMAAGLVKLAQEDPSFHFSRDEEI 1627
Query: 514 GEHIIAAAGELHLEICLKDLQDDFMNGAEITKSDPIVSFRETVLE----KSSHTVMSKSP 569
+ +I GELHLEI + L+ ++ E P V++RE++ + + H S
Sbjct: 1628NQTVIEGMGELHLEIIVDRLKREYK--VEANVGAPQVNYRESISKIHEARYVHKKQSGGQ 1801
Query: 570 NKHNRLYMEARPMEEG 585
+ + + PME G
Sbjct: 1802GQFADITVRFEPMEPG 1849
Score = 57.8 bits (138), Expect = 2e-08
Identities = 34/97 (35%), Positives = 51/97 (52%)
Frame = +1
Query: 698 ARRVFYAAMLTAKPRLLEPVYLVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNV 757
AR F + A PR+LEP+ VE+ PE LG + LN +RG + +P L V
Sbjct: 2047 ARGAFREGIRKAGPRMLEPIMKVEVVTPEEHLGDVIGDLNSRRGQINSFGDKPG-GLKVV 2223
Query: 758 KAYLPVIESFQFNESLRAQTGGQAFPQLVFDHWDMVP 794
+ +P+ E FQ+ +LR T G+A + +D+VP
Sbjct: 2224 DSLVPLAEMFQYVSTLRGMTKGRASYSMQLAMFDVVP 2334
>TC78337 similar to GP|9759359|dbj|BAB10014.1 GTP-binding protein LepA
homolog {Arabidopsis thaliana}, partial (40%)
Length = 1035
Score = 102 bits (255), Expect = 5e-22
Identities = 60/144 (41%), Positives = 80/144 (54%)
Frame = +1
Query: 6 NIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISL 65
NIRN S+IAH+DHGKSTL D L+ G + Q + + D E ERGITIK +
Sbjct: 256 NIRNFSIIAHIDHGKSTLADKLLQVTGTVPQREMKE-QFLDNMDLERERGITIKLQAARM 432
Query: 66 YYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCV 125
Y E Y +NLID+PGHVDFS EV+ +L +GAL+VVD +GV
Sbjct: 433 RYVF------------ENEPYCLNLIDTPGHVDFSYEVSRSLAACEGALLVVDASQGVEA 576
Query: 126 QTETVLRQALGERIKPVLTVNKMD 149
QT + AL ++ + +NK+D
Sbjct: 577 QTLANVYLALDNNLEIIPVLNKID 648
>TC77109 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein typA
(tyrosine phosphorylated protein A) {Arabidopsis
thaliana}, partial (27%)
Length = 672
Score = 99.4 bits (246), Expect = 5e-21
Identities = 59/145 (40%), Positives = 82/145 (55%)
Frame = +3
Query: 6 NIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISL 65
+IRN++++AHVDHGK+TL D+++ + R+ D+ E ERGITI S S+
Sbjct: 183 DIRNIAIVAHVDHGKTTLVDAMLKQTKVFRDNQTVQERIMDSNDLERERGITILSKNTSV 362
Query: 66 YYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCV 125
Y+ D K IN+ID+PGH DF EV L + +G L+VVD VEG
Sbjct: 363 TYK----DAK------------INIIDTPGHSDFGGEVERILNMVEGILLVVDSVEGPMP 494
Query: 126 QTETVLRQALGERIKPVLTVNKMDR 150
QT VL++AL V+ VNK+DR
Sbjct: 495 QTRFVLKKALEFGHAVVVVVNKIDR 569
>TC77108 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein typA
(tyrosine phosphorylated protein A) {Arabidopsis
thaliana}, partial (98%)
Length = 2589
Score = 99.4 bits (246), Expect = 5e-21
Identities = 59/145 (40%), Positives = 82/145 (55%)
Frame = +3
Query: 6 NIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISL 65
+IRN++++AHVDHGK+TL D+++ + R+ D+ E ERGITI S S+
Sbjct: 393 DIRNIAIVAHVDHGKTTLVDAMLKQTKVFRDNQTVQERIMDSNDLERERGITILSKNTSV 572
Query: 66 YYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCV 125
Y+ D K IN+ID+PGH DF EV L + +G L+VVD VEG
Sbjct: 573 TYK----DAK------------INIIDTPGHSDFGGEVERILNMVEGILLVVDSVEGPMP 704
Query: 126 QTETVLRQALGERIKPVLTVNKMDR 150
QT VL++AL V+ VNK+DR
Sbjct: 705 QTRFVLKKALEFGHAVVVVVNKIDR 779
Score = 29.6 bits (65), Expect = 5.0
Identities = 17/73 (23%), Positives = 34/73 (46%), Gaps = 1/73 (1%)
Frame = +2
Query: 719 LVEIQAPEHALGGIYSVLNQKRGHVFDEIQRPNTPLYNVKAY-LPVIESFQFNESLRAQT 777
+ ++ PE +G + +L Q+RG +FD +Q + N+ Y +P ++ +
Sbjct: 1682 IATVEVPEEHMGSVVELLGQRRGQMFD-MQGVGSEGTNLLKYKIPTRGLLGLRNAILTAS 1858
Query: 778 GGQAFPQLVFDHW 790
G A +FD +
Sbjct: 1859 RGTAILNTIFDSY 1897
>TC79024 similar to PIR|A84716|A84716 probable GTP-binding protein
[imported] - Arabidopsis thaliana, partial (67%)
Length = 1513
Score = 95.1 bits (235), Expect = 1e-19
Identities = 57/147 (38%), Positives = 85/147 (57%), Gaps = 1/147 (0%)
Frame = +2
Query: 5 HNIRNMSVIAHVDHGKSTLTDSLVAAAGI-IAQEVAGDVRMTDTRQDEAERGITIKSTGI 63
+ +RN++VIAHVDHGK+TL D L+ G I E R D+ E ERGITI S
Sbjct: 197 NRLRNVAVIAHVDHGKTTLMDRLLRQCGADIPHE-----RAMDSISLERERGITISSKVT 361
Query: 64 SLYYEMSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGV 123
S+ ++ ++ +N++D+PGH DF EV + + +GA++VVD EG
Sbjct: 362 SISWKDNE----------------LNMVDTPGHADFGGEVERVVGMVEGAVLVVDAGEGP 493
Query: 124 CVQTETVLRQALGERIKPVLTVNKMDR 150
QT+ VL +AL ++P+L +NK+DR
Sbjct: 494 LAQTKFVLAKALKYGLRPILLLNKVDR 574
>TC84431 similar to GP|10176983|dbj|BAB10215. GTP-binding membrane protein
LepA homolog {Arabidopsis thaliana}, partial (46%)
Length = 976
Score = 87.4 bits (215), Expect = 2e-17
Identities = 51/131 (38%), Positives = 78/131 (58%), Gaps = 1/131 (0%)
Frame = +3
Query: 21 STLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISLYYE-MSDGDLKNFKG 79
+TL D L+ G I + + G + D Q E ERGIT+K+ +++Y+ + +GD +FK
Sbjct: 3 TTLADRLLELTGTIKKGL-GQPQYLDKLQVERERGITVKAQTATMFYKNIINGD--DFKD 173
Query: 80 EREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQTETVLRQALGERI 139
+E + YL+NLID+PGHVDFS EV+ +L G L+VVD +GV QT A +
Sbjct: 174 GKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTVANFYLAFESNL 353
Query: 140 KPVLTVNKMDR 150
+ +NK+D+
Sbjct: 354 AIIPVINKIDQ 386
>TC90097 homologue to PIR|H86197|H86197 hypothetical protein [imported] -
Arabidopsis thaliana, partial (9%)
Length = 859
Score = 58.9 bits (141), Expect = 8e-09
Identities = 35/80 (43%), Positives = 47/80 (58%), Gaps = 12/80 (15%)
Frame = +2
Query: 762 PVIESFQFNESLRAQTGGQAFPQLVFDHWDMVPSDPLE------PGTPAAAR------VV 809
PVIESF F+ LR T GQAF Q VFDHW +VP DPL+ P PA + +V
Sbjct: 2 PVIESFGFDTDLRYHTQGQAFCQSVFDHWAIVPGDPLDKSIVLRPLEPAPIQHLAREFMV 181
Query: 810 EIRKKKGLKEQLIPLSEFED 829
+ R++KG+ E + + +F D
Sbjct: 182 KTRRRKGMSED-VSIGKFFD 238
>TC76989 homologue to SP|O24310|EFTU_PEA Elongation factor Tu chloroplast
precursor (EF-Tu). [Garden pea] {Pisum sativum}, partial
(92%)
Length = 1907
Score = 57.4 bits (137), Expect = 2e-08
Identities = 47/174 (27%), Positives = 73/174 (41%), Gaps = 1/174 (0%)
Frame = +3
Query: 9 NMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRQDEAERGITIKSTGISLYYE 68
N+ I HVDHGK+TLT +L A + D +E RGITI + +
Sbjct: 423 NIGTIGHVDHGKTTLTAALTMALASLGNSAPKKYDEIDAAPEERARGITINTATVEY--- 593
Query: 69 MSDGDLKNFKGEREGNKYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCVEGVCVQTE 128
E E Y +D PGH D+ + DGA++VV +G QT+
Sbjct: 594 -----------ETETRHYA--HVDCPGHADYVKNMITGAAQMDGAILVVSGADGPMPQTK 734
Query: 129 TVLRQALGERIKPVLT-VNKMDRCFLELHLDAEEAYSTIQRVIESVNVVMATYE 181
+ A + V+ +NK D+ +D EE ++ V ++++YE
Sbjct: 735 EHILLAKQVGVPSVVVFLNKQDQ------VDDEELLELVEL---EVRELLSSYE 869
>TC85417 similar to SP|Q39211|RP3A_ARATH DNA-directed RNA polymerase II 36
kDa polypeptide A (EC 2.7.7.6) (RNA polymerase II
subunit 3)., partial (64%)
Length = 855
Score = 50.8 bits (120), Expect = 2e-06
Identities = 22/41 (53%), Positives = 29/41 (70%)
Frame = +1
Query: 783 PQLVFDHWDMVPSDPLEPGTPAAARVVEIRKKKGLKEQLIP 823
P VFDHWD + SDPLE G+ AA V +I ++KG +EQ+ P
Sbjct: 733 PTRVFDHWDTMTSDPLEAGSQAAQLVTDIPQRKGTQEQMTP 855
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,604,265
Number of Sequences: 36976
Number of extensions: 289584
Number of successful extensions: 1362
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 1333
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1346
length of query: 831
length of database: 9,014,727
effective HSP length: 104
effective length of query: 727
effective length of database: 5,169,223
effective search space: 3758025121
effective search space used: 3758025121
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146817.8


