

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146807.2 + phase: 0
(309 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
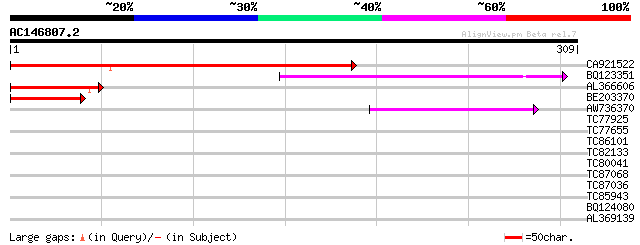
Score E
Sequences producing significant alignments: (bits) Value
CA921522 199 1e-51
BQ123351 102 2e-22
AL366606 78 4e-15
BE203370 60 1e-09
AW736370 44 8e-05
TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1 ... 30 1.2
TC77655 similar to PIR|T09217|T09217 protein sam2B - spinach, pa... 30 1.2
TC86101 similar to GP|15217283|gb|AAK92627.1 Putative leucine-ri... 28 3.4
TC82133 similar to PIR|G84597|G84597 probable XAP-5 protein [Hom... 28 3.4
TC80041 similar to GP|8778702|gb|AAF79710.1| T1N15.20 {Arabidops... 28 3.4
TC87068 similar to PIR|G96703|G96703 unknown protein 30164-3299... 28 3.4
TC87036 similar to GP|11138330|gb|AAG31326.1 putative serine/thr... 28 3.4
TC85943 similar to PIR|T07080|T07080 shock protein SRC2 - soybea... 27 7.6
BQ124080 weakly similar to GP|20197140|gb| predicted protein {Ar... 27 10.0
AL369139 weakly similar to GP|21554368|gb| unknown {Arabidopsis ... 27 10.0
>CA921522
Length = 767
Score = 199 bits (505), Expect = 1e-51
Identities = 98/191 (51%), Positives = 125/191 (65%), Gaps = 2/191 (1%)
Frame = +3
Query: 1 MNAIEIFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCVPLLYRWFISHLPSS--FHDNS 58
+ AI+IF S+NPVPTLL DTY +IH RT GRG ILCC LLYRW SHLP + F N
Sbjct: 186 IKAIQIFLSRNPVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSHLPRTPRFTTNP 365
Query: 59 ENWSYSQRIMALTPNEVVWITPTAQVKEIITGCGDFLNVPLLGTRGGINYNPELAMRQFG 118
EN +S+R+M+LTP EVVW II CG F NVPLLG GGI+YNP LA QFG
Sbjct: 366 ENLLWSKRLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISYNPTLARHQFG 545
Query: 119 FPMKAKPINLATSPEFFYYSNAPTGQREAFIGAWSKVCRKSVKHLGVRSGIAHEAYTQWV 178
+PM+ KP+++ ++ ++ TG RE + AW + R+ LG ++G +YTQWV
Sbjct: 546 YPMERKPLSIYLENVYYLNADDSTGMREHVVRAWHTIRRRDKDQLGKKTGAISSSYTQWV 725
Query: 179 INRAEEIGMPY 189
I+RA +IGMPY
Sbjct: 726 IDRAVQIGMPY 758
>BQ123351
Length = 725
Score = 102 bits (254), Expect = 2e-22
Identities = 60/157 (38%), Positives = 93/157 (59%)
Frame = +2
Query: 148 FIGAWSKVCRKSVKHLGVRSGIAHEAYTQWVINRAEEIGMPYPAMRYVSASAPSIPLPLP 207
FI AW V ++ LG RS + +E+YT+WVI+RA + G+P+P R S++ S LP+
Sbjct: 8 FIQAWRAVHKREKGQLGRRS*LVYESYTKWVIDRAAKNGIPFPLQRLPSSTTLSPSLPMT 187
Query: 208 PTTQGMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLMLKKELVKVRREL 267
T+ Q LA +REK W+ RY EAE+ I TL G+ EQK HE L +++++++ L
Sbjct: 188 SKTKEEAQYLLAEMTREKDTWRIRYMEAESEIGTLRGQVEQKDHELLKMRQQMIERDDLL 367
Query: 268 EEKDELLMRDSKRARGRRNFYARYCGSDSESESEDHP 304
+EKD+LL + + + R + + G DS+ E P
Sbjct: 368 QEKDKLLKKHITK-KQRMDSMDLFDGPDSDFED*SFP 475
>AL366606
Length = 393
Score = 78.2 bits (191), Expect = 4e-15
Identities = 41/57 (71%), Positives = 43/57 (74%), Gaps = 6/57 (10%)
Frame = -1
Query: 1 MNAIEIFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCVPLL------YRWFISHLP 51
MNAIE+FHSKNPVPTLLADTYHAIHDRTLKGRGYIL L +R F S LP
Sbjct: 318 MNAIEVFHSKNPVPTLLADTYHAIHDRTLKGRGYILWLHILFSNRVVYFRTFRSSLP 148
>BE203370
Length = 454
Score = 59.7 bits (143), Expect = 1e-09
Identities = 27/41 (65%), Positives = 33/41 (79%)
Frame = +1
Query: 1 MNAIEIFHSKNPVPTLLADTYHAIHDRTLKGRGYILCCVPL 41
++AI++F +KNPVPTLLAD YH+IHDR GRG IL C PL
Sbjct: 331 IHAIQMFLTKNPVPTLLADMYHSIHDRNQVGRGAILGCAPL 453
>AW736370
Length = 488
Score = 43.9 bits (102), Expect = 8e-05
Identities = 26/92 (28%), Positives = 43/92 (46%)
Frame = -2
Query: 197 ASAPSIPLPLPPTTQGMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLML 256
A P+ PLP+ T+ YQE L RE WK Y + + T+ G EQ+ +++
Sbjct: 487 AITPAPPLPMTFDTEKEYQERLTEAEREIHRWKREYQKKDQDYETVMGLLEQEAYDSR*K 308
Query: 257 KKELVKVRRELEEKDELLMRDSKRARGRRNFY 288
+ K+ ++E D L R R + R + +
Sbjct: 307 DVIIAKLNERIKENDAALDRIPGRKKRRMDLF 212
>TC77925 similar to GP|18700163|gb|AAL77693.1 AT4g02720/T10P11_1
{Arabidopsis thaliana}, partial (47%)
Length = 1538
Score = 30.0 bits (66), Expect = 1.2
Identities = 17/44 (38%), Positives = 24/44 (53%)
Frame = +3
Query: 258 KELVKVRRELEEKDELLMRDSKRARGRRNFYARYCGSDSESESE 301
KE+ + + E +E DS+ + RR Y + SDSESESE
Sbjct: 522 KEIKQKAKSESESEESKESDSELRKKRRKSYKKSRESDSESESE 653
>TC77655 similar to PIR|T09217|T09217 protein sam2B - spinach, partial (64%)
Length = 1341
Score = 30.0 bits (66), Expect = 1.2
Identities = 16/48 (33%), Positives = 29/48 (60%), Gaps = 3/48 (6%)
Frame = +3
Query: 243 DGKDEQKTHENLMLKKELVKVRRELEEK---DELLMRDSKRARGRRNF 287
DG++E + + + +KKEL KV +E E+ +L+ K+A GR ++
Sbjct: 468 DGEEESEEEKRMRVKKELEKVAKEQAERRATAQLMYDLGKKAYGRGSY 611
>TC86101 similar to GP|15217283|gb|AAK92627.1 Putative leucine-rich repeat
transmembrane protein kinase 1 {Oryza sativa}, partial
(64%)
Length = 2629
Score = 28.5 bits (62), Expect = 3.4
Identities = 18/71 (25%), Positives = 33/71 (46%)
Frame = +1
Query: 169 IAHEAYTQWVINRAEEIGMPYPAMRYVSASAPSIPLPLPPTTQGMYQEHLAMESREKQMW 228
I + +T W+ + + I + + S AP P PP T+ L ++ +Q+
Sbjct: 925 IENNHFTGWIPEQLKNINLQKNGNSWSSGPAPPPPPGTPPVTKTKVITSLVVDIAPRQI- 1101
Query: 229 KARYNEAENLI 239
A+ EA+NL+
Sbjct: 1102LAQVMEAKNLV 1134
>TC82133 similar to PIR|G84597|G84597 probable XAP-5 protein [Homo sapiens]
[imported] - Arabidopsis thaliana, partial (60%)
Length = 926
Score = 28.5 bits (62), Expect = 3.4
Identities = 18/84 (21%), Positives = 37/84 (43%), Gaps = 3/84 (3%)
Frame = -2
Query: 134 FFYYSNAPTGQREAFIGAWSK---VCRKSVKHLGVRSGIAHEAYTQWVINRAEEIGMPYP 190
F YS+ T +F+ A S+ V + + G +A ++ W+ R+ +
Sbjct: 400 FSTYSSRVTSPTVSFLKAVSRISEVLEPNWRRPGWPLEVADLLFSSWIFRRSASLCFSSR 221
Query: 191 AMRYVSASAPSIPLPLPPTTQGMY 214
+R S + P+ P P+P + ++
Sbjct: 220 RIRTASWAVPTYPSPIPDISLALF 149
>TC80041 similar to GP|8778702|gb|AAF79710.1| T1N15.20 {Arabidopsis
thaliana}, partial (7%)
Length = 1621
Score = 28.5 bits (62), Expect = 3.4
Identities = 34/117 (29%), Positives = 49/117 (41%), Gaps = 12/117 (10%)
Frame = +3
Query: 200 PSIPLPLPPT-TQGMYQEHLAMESREKQMWKARYNEAENLIMT-----LDGKDEQKT--- 250
PS L LP TQG+ +L +ESR +++ ENL+++ G QKT
Sbjct: 705 PSSVLSLPQERTQGILN-NLRVESRFREVAGNGKQRDENLVLSGCHFDNGGDQNQKTVSD 881
Query: 251 HENLMLKKELVKVRRELEEKDELLMRDSKRARGRRNFYARYCGSDSESESED---HP 304
ENL L++++V+ + CG D ESE ED HP
Sbjct: 882 RENLSLRQDIVRPVEAI------------------------CGGDLESEMEDQQNHP 980
>TC87068 similar to PIR|G96703|G96703 unknown protein 30164-32998
[imported] - Arabidopsis thaliana, partial (24%)
Length = 711
Score = 28.5 bits (62), Expect = 3.4
Identities = 17/68 (25%), Positives = 30/68 (44%)
Frame = +2
Query: 207 PPTTQGMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLMLKKELVKVRRE 266
PP++ + E WK ++ N + +D KD +L+KE + +R+
Sbjct: 185 PPSSSFNQSSERTKDKEENNNWKGSLDDKPNDLPLVDTKDNVPGVLYDLLQKEGLGLRKA 364
Query: 267 LEEKDELL 274
EKD+ L
Sbjct: 365 GHEKDQSL 388
>TC87036 similar to GP|11138330|gb|AAG31326.1 putative serine/threonine
kinase GDBrPK {Vitis vinifera}, partial (66%)
Length = 1212
Score = 28.5 bits (62), Expect = 3.4
Identities = 9/18 (50%), Positives = 13/18 (72%)
Frame = +2
Query: 45 WFISHLPSSFHDNSENWS 62
WF+ +LP F ++ ENWS
Sbjct: 692 WFLKNLPLEFMEDGENWS 745
>TC85943 similar to PIR|T07080|T07080 shock protein SRC2 - soybean, partial
(74%)
Length = 1279
Score = 27.3 bits (59), Expect = 7.6
Identities = 14/45 (31%), Positives = 27/45 (59%)
Frame = -1
Query: 201 SIPLPLPPTTQGMYQEHLAMESREKQMWKARYNEAENLIMTLDGK 245
++P+ + P T + +++ SRE++ W AR E+L++ L GK
Sbjct: 409 TLPIKVSPAT---VRSEMSLTSRERRFWLAR----ESLMVNLTGK 296
>BQ124080 weakly similar to GP|20197140|gb| predicted protein {Arabidopsis
thaliana}, partial (22%)
Length = 660
Score = 26.9 bits (58), Expect = 10.0
Identities = 12/29 (41%), Positives = 17/29 (58%)
Frame = +1
Query: 176 QWVINRAEEIGMPYPAMRYVSASAPSIPL 204
QW+I+ I PYP ++ S+PS PL
Sbjct: 322 QWIIHACVSIWWPYP---FLDLSSPSAPL 399
>AL369139 weakly similar to GP|21554368|gb| unknown {Arabidopsis thaliana},
partial (11%)
Length = 388
Score = 26.9 bits (58), Expect = 10.0
Identities = 24/69 (34%), Positives = 31/69 (44%), Gaps = 12/69 (17%)
Frame = +3
Query: 245 KDEQKTHENLMLKKEL-------VKVRRELEEKDELLMRDSKRARGRRNFYARY-----C 292
+D +KT EN + E K RE E D R KR + RR + +
Sbjct: 66 RDSRKTRENSDIDTETDRKHISSKKKSRESSESDSDKRRRRKR-KSRRKYSSESDSESDS 242
Query: 293 GSDSESESE 301
GSDSESES+
Sbjct: 243 GSDSESESD 269
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.134 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,331,854
Number of Sequences: 36976
Number of extensions: 148398
Number of successful extensions: 898
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 889
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 897
length of query: 309
length of database: 9,014,727
effective HSP length: 96
effective length of query: 213
effective length of database: 5,465,031
effective search space: 1164051603
effective search space used: 1164051603
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146807.2


