

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146806.4 + phase: 0 /pseudo
(796 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
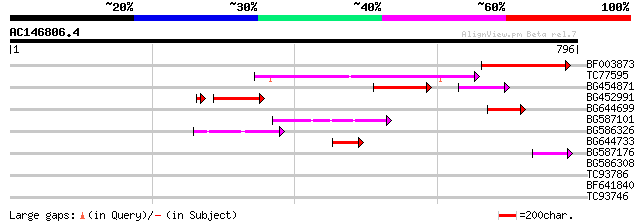
Score E
Sequences producing significant alignments: (bits) Value
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 167 2e-41
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 164 1e-40
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 75 1e-17
BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, pa... 76 6e-16
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 65 8e-11
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 57 2e-08
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 56 6e-08
BG644733 weakly similar to GP|15289942|db putative polyprotein {... 47 2e-05
BG587176 weakly similar to PIR|G84493|G84 probable retroelement ... 47 4e-05
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 36 0.066
TC93786 similar to PIR|F84542|F84542 nodulin-like protein [impor... 31 1.6
BF641840 similar to GP|14334538|gb unknown protein {Arabidopsis ... 30 2.8
TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical... 29 6.2
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 167 bits (422), Expect = 2e-41
Identities = 81/125 (64%), Positives = 98/125 (77%)
Frame = +2
Query: 663 GVGHALKCRKLTPRFVGPFDVIEKVGVVAYRIALPPSLSNLHNVFHVPKLRKYVHDASHV 722
GVG ALK +KLT RF+GP+ + E+VG VAYR+ LPP L NLH+VFHV +LRKYV D SHV
Sbjct: 2 GVGRALKSKKLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQLRKYVPDPSHV 181
Query: 723 IRVDDLEVRDNLTVETWPVRIEDRELKRLRGKEIVLVKVIWVGPTGESATWEPESGMMVS 782
I+ DD++VRDNLTVET PVRI+DR++K LRGKEI LV+V+W GES TWE ES M+ S
Sbjct: 182 IQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWDRANGESLTWELESKMVES 361
Query: 783 YRSCF 787
Y F
Sbjct: 362 YPELF 376
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial
(14%)
Length = 1708
Score = 164 bits (415), Expect = 1e-40
Identities = 111/330 (33%), Positives = 163/330 (48%), Gaps = 14/330 (4%)
Frame = +2
Query: 344 RLDENDVLMFRDRVCVPDVLD-------LKRQILDEGHRSSLSIHPGATKMYQDLKRLFW 396
+LD L FR R+ VP D L+ +++ E H S+ + HPG + + R F+
Sbjct: 83 QLDSLKRLTFRGRIWVPGSDDEESPLNELRTKLVQESHDSTAAGHPGRNGTLEIVSRKFF 262
Query: 397 WPGMKKEIAEFVYACLVCQKSMIEHQRPSGLMQPLFVPEWTWDSISMDFVGALPKT-SKS 455
WPG + + FV C VC I Q G ++PL VP +SMDF+ +LP T +
Sbjct: 263 WPGQSQTVRRFVRNCDVCGGIHIWRQAKRGFLKPLPVPNRLHSDLSMDFITSLPPTRGRG 442
Query: 456 FDTIWVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQIVRLHGIPSSIVSDRDPRFTSNFW 515
+WVIVDRL+KS + T M A+ ++ R HG+P SIVSDR + FW
Sbjct: 443 SQYLWVIVDRLSKSVTLEEMDT-MEAEACAQRFLSCHYRFHGMPQSIVSDRGSNWVGRFW 619
Query: 516 ESLQAALGTKLSLSSAYHPQTDGQTERTIQSLEDLLRACVLEQGVSWDECLPLIEFTYNN 575
G LS++YHPQTDG TER Q ++ +LRA V +W + LP ++ N
Sbjct: 620 REFCRLTGVTQLLSTSYHPQTDGGTERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRN 799
Query: 576 SFHSSIRMAPFEALYGRRCRTPLCWFES-----GESAMLAPEVVQETTEKVKMIQEKMKA 630
+SSI PF +G P+ E E A +V+ + IQ ++ A
Sbjct: 800 RHNSSIGATPFFVEHGYHV-DPIPTVEDTGGVVSEGEAAAQLLVKRMKDVTGFIQAEIVA 976
Query: 631 SQST*KSYHDKRRKDIE-FQVGDHVFLWVN 659
+Q ++ +KRR + +QVGD V+L V+
Sbjct: 977 AQQRSEASANKRRCPADRYQVGDKVWLNVS 1066
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 75.5 bits (184), Expect(2) = 1e-17
Identities = 36/81 (44%), Positives = 49/81 (60%)
Frame = +2
Query: 512 SNFWESLQAALGTKLSLSSAYHPQTDGQTERTIQSLEDLLRACVLEQGVSWDECLPLIEF 571
SNFW+ L GT L++SSAYHP +DGQ+E + E LR + + W + P E+
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 572 TYNNSFHSSIRMAPFEALYGR 592
YN S++ S M PF+ALYGR
Sbjct: 212 WYNTSYNISAAMTPFKALYGR 274
Score = 33.1 bits (74), Expect(2) = 1e-17
Identities = 25/73 (34%), Positives = 35/73 (47%), Gaps = 2/73 (2%)
Frame = +1
Query: 631 SQST*KSYHDKRRKDIEFQVGDHVFLWVNPVTGVGHAL-KCRKL-TPRFVGPFDVIEKVG 688
+Q T K DK+R+ EFQ+G+HV + + P AL K +K +P F V
Sbjct: 388 AQQTMKHQADKKRRHFEFQLGEHVLVKLQPYQQSSVALRKYQKFGSPNFGSLLTVCSL*V 567
Query: 689 VVAYRIALPPSLS 701
A+ PP LS
Sbjct: 568 ESAFHCKSPPYLS 606
>BG452991 PIR|A25875|A25 histone H4 - Tetrahymena thermophila, partial (33%)
Length = 560
Score = 75.9 bits (185), Expect(2) = 6e-16
Identities = 36/72 (50%), Positives = 52/72 (72%)
Frame = +3
Query: 286 IEQFGELSLVSELTPDGVRLGMLKLTSNILEEIKNGQKEDLELVDRVTLVNQGKGGDFRL 345
+EQF +LSLV E++P V+LGMLK+ + L+ IK QK D++LVD + NQ + GDF++
Sbjct: 69 LEQFRDLSLVCEVSPQSVKLGMLKINNEFLDSIKEAQKVDVKLVDLMFGNNQTEDGDFKV 248
Query: 346 DENDVLMFRDRV 357
D+ VL FRDR+
Sbjct: 249 DDQGVLQFRDRI 284
Score = 26.9 bits (58), Expect(2) = 6e-16
Identities = 12/13 (92%), Positives = 13/13 (99%)
Frame = +1
Query: 263 VVADVLSRKTLHM 275
VVADVLSRKTLH+
Sbjct: 1 VVADVLSRKTLHV 39
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 65.5 bits (158), Expect = 8e-11
Identities = 27/53 (50%), Positives = 41/53 (76%)
Frame = +2
Query: 672 KLTPRFVGPFDVIEKVGVVAYRIALPPSLSNLHNVFHVPKLRKYVHDASHVIR 724
KL+ R++GPF+VI+++G VAY +ALPP LS +H VFHV ++Y D +++IR
Sbjct: 65 KLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVSMFKRYHGDGNYIIR 223
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 57.4 bits (137), Expect = 2e-08
Identities = 44/168 (26%), Positives = 79/168 (46%), Gaps = 1/168 (0%)
Frame = +2
Query: 369 ILDEGHRSSLSIHPGATKMYQDLKRL-FWWPGMKKEIAEFVYACLVCQKSMIEHQRPSGL 427
IL H S+ + H +K +++ FWWP M K+ F+ C CQ+ R + +
Sbjct: 104 ILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPCQRQGNIS*R-NEM 280
Query: 428 MQPLFVPEWTWDSISMDFVGALPKTSKSFDTIWVIVDRLTKSTHFVPIKTSMSIARLAEI 487
Q + +D +DF+G P +S + I V VD ++K + T+ + + ++
Sbjct: 281 PQNFILEVEVFDVWGIDFMGPFP-SSYNNKYILVAVDYVSKWVEAIASPTNDATV-VVKM 454
Query: 488 YIEQIVRLHGIPSSIVSDRDPRFTSNFWESLQAALGTKLSLSSAYHPQ 535
+ I G+P ++SD F + +E L G + +++AYHPQ
Sbjct: 455 FKSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQ 598
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 55.8 bits (133), Expect = 6e-08
Identities = 36/130 (27%), Positives = 69/130 (52%), Gaps = 1/130 (0%)
Frame = +2
Query: 258 PGKANVVADVLSRKTLHMYALMVKELELIEQFGELSLVSELTPDGVRLGMLKLT-SNILE 316
PGKAN+VAD LSR+ + + A +E + ++ ++ LT LG+ + +++
Sbjct: 353 PGKANLVADALSRRRVDVSA--EREADDLDGMVRALRLNVLTKATESLGLEAVNQADLFT 526
Query: 317 EIKNGQKEDLELVDRVTLVNQGKGGDFRLDENDVLMFRDRVCVPDVLDLKRQILDEGHRS 376
I+ Q +D + + V Q +++ ++ ++ R+ VP+ LK +I+ E H+S
Sbjct: 527 RIRLAQGQD----ENLQKVAQNDRTEYQTAKDGTILVNGRISVPNDRSLKEEIMSEAHKS 694
Query: 377 SLSIHPGATK 386
S+HPGA +
Sbjct: 695 RFSVHPGAPR 724
>BG644733 weakly similar to GP|15289942|db putative polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (1%)
Length = 174
Score = 47.4 bits (111), Expect = 2e-05
Identities = 23/43 (53%), Positives = 32/43 (73%)
Frame = -3
Query: 454 KSFDTIWVIVDRLTKSTHFVPIKTSMSIARLAEIYIEQIVRLH 496
KS+++I V+VDRLTKST F+P KTS S A I +++IV +H
Sbjct: 130 KSYESI*VVVDRLTKSTLFIPFKTSYSAK*YARILLDEIVCIH 2
>BG587176 weakly similar to PIR|G84493|G84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(1%)
Length = 729
Score = 46.6 bits (109), Expect = 4e-05
Identities = 25/56 (44%), Positives = 31/56 (54%)
Frame = -1
Query: 734 LTVETWPVRIEDRELKRLRGKEIVLVKVIWVGPTGESATWEPESGMMVSYRSCFLQ 789
L +ET PVRI DR K +R K I +VK++W E TWE E+ M Y F Q
Sbjct: 717 LDLETRPVRILDRMEKAMRKKPIQMVKIVWDCSGREEITWETEARMKADYPEWFNQ 550
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 686
Score = 35.8 bits (81), Expect = 0.066
Identities = 22/70 (31%), Positives = 37/70 (52%), Gaps = 1/70 (1%)
Frame = -2
Query: 496 HGIPSSIVSDRDPRFTSNFWESLQAALGTKLSLSSAYHPQTDGQTERTIQSLED-LLRAC 554
HG+P IV+D F SN + +L+ +S +PQ++GQ E + + + D L +
Sbjct: 685 HGLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRL 506
Query: 555 VLEQGVSWDE 564
L++G DE
Sbjct: 505 DLKKGCWADE 476
>TC93786 similar to PIR|F84542|F84542 nodulin-like protein [imported] -
Arabidopsis thaliana, partial (26%)
Length = 554
Score = 31.2 bits (69), Expect = 1.6
Identities = 15/38 (39%), Positives = 21/38 (54%), Gaps = 1/38 (2%)
Frame = -1
Query: 668 LKCRKLTPRFVGPFDV-IEKVGVVAYRIALPPSLSNLH 704
L+CRKL PRF G F + + V + + PP N+H
Sbjct: 497 LECRKLEPRFFGKFLMQVTSTAVFIHVLLFPPIHKNIH 384
>BF641840 similar to GP|14334538|gb unknown protein {Arabidopsis thaliana},
partial (26%)
Length = 654
Score = 30.4 bits (67), Expect = 2.8
Identities = 23/72 (31%), Positives = 39/72 (53%), Gaps = 6/72 (8%)
Frame = +1
Query: 463 VDRLTKSTHFVPIKTSMS-----IARLAEIYIEQIVRLHGIPS-SIVSDRDPRFTSNFWE 516
V R+ KS +P+ S+S + L +I+++ + L+G P IVSDR+P++ S W
Sbjct: 319 VSRVCKS---IPLSPSVSREERHSSSLKDIHVDVVCILNGKPVWIIVSDRNPKYIS--WN 483
Query: 517 SLQAALGTKLSL 528
+ G KL +
Sbjct: 484 ECHKSKGLKLRI 519
>TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical
protein~similar to gag-pol polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (4%)
Length = 1019
Score = 29.3 bits (64), Expect = 6.2
Identities = 14/47 (29%), Positives = 28/47 (58%)
Frame = +2
Query: 427 LMQPLFVPEWTWDSISMDFVGALPKTSKSFDTIWVIVDRLTKSTHFV 473
L++P VP+ W+ +++DF L T + D+ V+ D+ ++ HF+
Sbjct: 464 LLRP--VPKPPWEDVTIDFSLGLL*TQQLKDSKMVVGDKFSRMAHFI 598
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.340 0.151 0.500
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,951,544
Number of Sequences: 36976
Number of extensions: 306217
Number of successful extensions: 1939
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 1125
Number of HSP's successfully gapped in prelim test: 92
Number of HSP's that attempted gapping in prelim test: 796
Number of HSP's gapped (non-prelim): 1241
length of query: 796
length of database: 9,014,727
effective HSP length: 104
effective length of query: 692
effective length of database: 5,169,223
effective search space: 3577102316
effective search space used: 3577102316
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146806.4


