

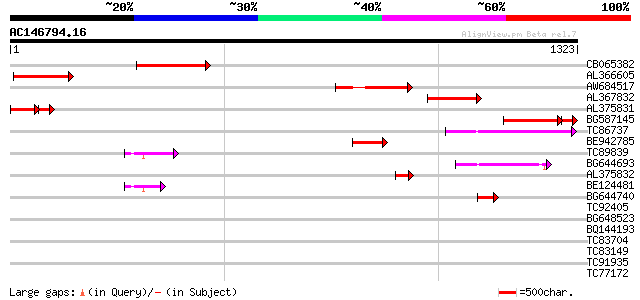
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146794.16 + phase: 0 /pseudo/partial
(1323 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fra... 305 8e-83
AL366605 292 5e-79
AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thalia... 252 8e-67
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 240 2e-63
AL375831 142 8e-51
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 152 8e-42
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 159 5e-39
BE942785 157 2e-38
TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC ... 91 4e-18
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 87 4e-17
AL375832 73 8e-13
BE124481 72 2e-12
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 49 2e-05
TC92405 similar to GP|11994325|dbj|BAB02284. gene_id:MLM24.13~un... 38 0.023
BG648523 similar to GP|9955540|emb| putative protein {Arabidopsi... 38 0.030
BQ144193 weakly similar to GP|14026007|dbj putative transporter ... 37 0.066
TC83704 similar to PIR|T05005|T05005 hypothetical protein T19P19... 35 0.19
TC83149 similar to GP|13561982|gb|AAK30594.1 flagelliform silk p... 34 0.43
TC91935 34 0.43
TC77172 similar to GP|20466472|gb|AAM20553.1 putative protein {A... 33 0.56
>CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fragment),
partial (9%)
Length = 624
Score = 305 bits (781), Expect = 8e-83
Identities = 147/172 (85%), Positives = 153/172 (88%)
Frame = -2
Query: 296 PVQQQQQNQQARPTFPPIPMLYAELLPTLLHRGHCTTRQGKPPPDPLPPRFRSDLKCDFH 355
PV QQ+Q QQARPTFP IPMLYAE LPTLL RGHCT RQGKPPPDPLPPRFRSDLKCDFH
Sbjct: 518 PVHQQRQ*QQARPTFPLIPMLYAE*LPTLLLRGHCTIRQGKPPPDPLPPRFRSDLKCDFH 339
Query: 356 QGALGHDVEGCYALKYIVKKLIDQGKLTFENNVPHVLDNPLPNHAAVNMIEVCEEAPRLD 415
QGALGHDVEGCYALK+IVKKLI+QGKLTFENNVPHVLDNPLPNHAAVNMIEV EEAP LD
Sbjct: 338 QGALGHDVEGCYALKHIVKKLINQGKLTFENNVPHVLDNPLPNHAAVNMIEVYEEAPGLD 159
Query: 416 VRNVATPLVPLHIKLCKASLFSHDHAKCLGCLRNPLGCYTVQDDIQGLMNDN 467
VRNV TPLVPLHIKLC+ASLF HDHA C NPLGC VQ+DIQ LMN+N
Sbjct: 158 VRNVTTPLVPLHIKLCQASLFDHDHANCQE*FYNPLGCCVVQNDIQSLMNNN 3
>AL366605
Length = 422
Score = 292 bits (748), Expect = 5e-79
Identities = 139/140 (99%), Positives = 139/140 (99%)
Frame = -3
Query: 10 HEIKANRGNADSFKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKD 69
HEIKANRGNADSFKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKD
Sbjct: 420 HEIKANRGNADSFKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKYVRKMGNYKD 241
Query: 70 NDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELAVAFKSHYGFNTRLKPNREFLRSLS 129
NDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELA AFKSHYGFNTRLKPNREFLRSLS
Sbjct: 240 NDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDELAAAFKSHYGFNTRLKPNREFLRSLS 61
Query: 130 QKKEESFREYAQRWRGAAAR 149
QKKEESFREYAQRWRGAAAR
Sbjct: 60 QKKEESFREYAQRWRGAAAR 1
>AW684517 similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 488
Score = 252 bits (643), Expect = 8e-67
Identities = 128/180 (71%), Positives = 140/180 (77%)
Frame = +1
Query: 761 ITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFAKSGKLVTIHGEEAYLVSQLSSF 820
ITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKF K+G
Sbjct: 1 ITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNG------------------- 123
Query: 821 SCIEAGSAEGTAFQGLTVEGTEPKRDGTAMASLKDAQRAVQEGQAAGWGRLIQLRENKHK 880
SAEGTAFQGL++EG EPK+ G AMASLKDAQ+AVQEGQAA WG+LIQL ENK K
Sbjct: 124 ------SAEGTAFQGLSMEGAEPKKVGAAMASLKDAQKAVQEGQAADWGKLIQLCENKRK 285
Query: 881 EGLGFSPTSGVSTGAFYSAGFVNAITEEATGFGPRPVFVIPGGIARDWDAIDIPSIMHVS 940
EGL FSPTSGVSTG F+SAGFVN + EE F PRP+FVIPGGIA+DWDA+D+PSIMHVS
Sbjct: 286 EGLRFSPTSGVSTGTFHSAGFVNTLAEEVARFVPRPLFVIPGGIAKDWDAVDVPSIMHVS 465
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 240 bits (613), Expect = 2e-63
Identities = 118/128 (92%), Positives = 122/128 (95%)
Frame = -1
Query: 974 QEKKAIQPH*EEIELINIGTEENKREIKIGAALEEGVKKKIIQLLREYPDIFAWSYEDMP 1033
QE+KAIQPH EEIELIN+GTEENKREIK+GAALEEGVK+KI QLLREY DIFA SYEDMP
Sbjct: 384 QERKAIQPHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLLREYLDIFACSYEDMP 205
Query: 1034 GLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANI 1093
GLDP IVEHRIPTKPECPPVR KLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANI
Sbjct: 204 GLDPKIVEHRIPTKPECPPVR*KLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANI 25
Query: 1094 VPVPKKDG 1101
VPVPKKDG
Sbjct: 24 VPVPKKDG 1
>AL375831
Length = 467
Score = 142 bits (359), Expect(2) = 8e-51
Identities = 67/69 (97%), Positives = 68/69 (98%)
Frame = +2
Query: 1 MEELAKELRHEIKANRGNADSFKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKY 60
MEELAKELRHEI+ANRGNADS KTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKY
Sbjct: 158 MEELAKELRHEIQANRGNADSVKTQDLCLVSKVDVPKKFKIPDFDRYNGLTCPQNHIIKY 337
Query: 61 VRKMGNYKD 69
VRKMGNYKD
Sbjct: 338 VRKMGNYKD 364
Score = 77.8 bits (190), Expect(2) = 8e-51
Identities = 35/36 (97%), Positives = 35/36 (97%)
Frame = +3
Query: 68 KDNDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDE 103
K NDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDE
Sbjct: 360 KTNDSLMIHCFQDSLMEDAAEWYTSLSKNDIHTFDE 467
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 152 bits (383), Expect(2) = 8e-42
Identities = 70/136 (51%), Positives = 101/136 (73%)
Frame = +2
Query: 1153 MSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVK 1212
M P+D EKT+FIT GT+CYKVMPFGL NAG+TYQR + +F D + +EVY+DDM+VK
Sbjct: 2 MHPDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVK 181
Query: 1213 STDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIR 1272
S H+ +L + F+ L +Y ++LNP KCTFGV SG+ LG+IV+Q+GIEV+P ++ AI
Sbjct: 182 SLRATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAIL 361
Query: 1273 EMPAPQTQKQVRGFLG 1288
++P+P+ ++V+ G
Sbjct: 362 DLPSPKNSREVQRLTG 409
Score = 38.5 bits (88), Expect(2) = 8e-42
Identities = 16/36 (44%), Positives = 24/36 (66%)
Frame = +3
Query: 1288 GRLNYISRFISHMTATCGPIFKLLRKNQPIVWNDEC 1323
GR+ ++RFIS T C P +KLL N+ VW+++C
Sbjct: 408 GRIAALNRFISRSTDKCLPFYKLLCGNKRFVWDEKC 515
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 159 bits (403), Expect = 5e-39
Identities = 101/313 (32%), Positives = 163/313 (51%), Gaps = 7/313 (2%)
Frame = +1
Query: 1017 LLREYPDIFAWSYEDMPGLDPMIVEHRIPTKPEC----PPVRQ-KLRRTHPDMALKIKSE 1071
+L E+PD+F +++H IP P+ PP+ L L +K
Sbjct: 343 VLEEFPDLFNPEKAYQVPASRGLLDHAIPLIPDKDGNDPPLPWGPLYGMSRQELLVLKKT 522
Query: 1072 VQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASLKDNFPLPHIDVLVD 1131
++ +D GF+ A ++ V K G +R CVD+R LN + KD +PLP I +
Sbjct: 523 LEDLLDKGFIKASGSAAG-APVLFVRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLR 699
Query: 1132 NTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMT 1191
A ++ F+ +D + ++++++ ED+EKT+F T +G F + V PFGL A AT+QR +
Sbjct: 700 RVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYIN 879
Query: 1192 TLFHDMIHKEVEVYVDDMIVKST-DEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGK 1250
H+ + V Y+DD+++ +T ++ H + ++ RL L L+P KC F V + K
Sbjct: 880 KTLHEFLDDFVTAYIDDVLIYTTGSKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVK 1059
Query: 1251 LLGFIVSQ-KGIEVDPDKVRAIREMPAPQTQKQVRGFLGRLNYISRFISHMTATCGPIFK 1309
+GFI++ KG+ DP K+ AIR+ P + K R FLG NY FI + P+ +
Sbjct: 1060 YVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTR 1239
Query: 1310 LLRKNQPIVWNDE 1322
L RK+ P W E
Sbjct: 1240 LTRKDFPFRWGAE 1278
>BE942785
Length = 460
Score = 157 bits (398), Expect = 2e-38
Identities = 78/82 (95%), Positives = 80/82 (97%)
Frame = -2
Query: 800 SGKLVTIHGEEAYLVSQLSSFSCIEAGSAEGTAFQGLTVEGTEPKRDGTAMASLKDAQRA 859
SG LVTIHGEEAYL+SQLSSFSCIEAGSAEGTAFQGLTVEGTEPKRDGTAMASLKDAQRA
Sbjct: 378 SGXLVTIHGEEAYLISQLSSFSCIEAGSAEGTAFQGLTVEGTEPKRDGTAMASLKDAQRA 199
Query: 860 VQEGQAAGWGRLIQLRENKHKE 881
VQE QAAGWGRLIQLRENKHK+
Sbjct: 198 VQESQAAGWGRLIQLRENKHKD 133
>TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC 4.6.1.1)
(ATP pyrophosphate-lyase) (Adenylyl cyclase). [Yeast],
partial (0%)
Length = 665
Score = 90.5 bits (223), Expect = 4e-18
Identities = 56/137 (40%), Positives = 69/137 (49%), Gaps = 11/137 (8%)
Frame = +3
Query: 267 PPFYHQYPLPPGQPQVPVNAIAQQMKQQLPVQQQQQNQQARPTFP-----------PIPM 315
PP Q P P QP P Q+ +QQ P Q QNQ + P PIP+
Sbjct: 228 PPLVQQRPQSPYQPMCP-----QKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPV 392
Query: 316 LYAELLPTLLHRGHCTTRQGKPPPDPLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKK 375
YA+LLP LL + T P+ LPP +R DL C FHQGA GHD E CY LK V+K
Sbjct: 393 KYADLLPILLKKNLIQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQK 572
Query: 376 LIDQGKLTFENNVPHVL 392
LI+ +F++ VL
Sbjct: 573 LIENNVWSFDDQDIKVL 623
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 87.0 bits (214), Expect = 4e-17
Identities = 65/230 (28%), Positives = 110/230 (47%), Gaps = 5/230 (2%)
Frame = +2
Query: 1040 VEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKK 1099
++ I P P+ R +P +K +++ ++ GF+ YP V ++ + KK
Sbjct: 35 IDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVV-VLFLKKK 211
Query: 1100 DGKVRMCVDFRDLNKASLKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDRE 1159
DG +RM +D+ LN ++K +PLP ID L DN SK F +D G +Q ++ ED
Sbjct: 212 DGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVIGEDVP 391
Query: 1160 KTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSTDEEQH 1219
KT+F +G + VM FG N + M +F D + V V+ +D+++ S +E +H
Sbjct: 392 KTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSKNENEH 571
Query: 1220 VEYLTKMFERLRKYKLRLNPNKCTFG-----VRSGKLLGFIVSQKGIEVD 1264
+L + L+ L C V G ++S +G++VD
Sbjct: 572 ENHLRLALKVLKDIGL------CQISYV*ILVEVGFFSLHVISGEGLKVD 703
>AL375832
Length = 535
Score = 72.8 bits (177), Expect = 8e-13
Identities = 34/41 (82%), Positives = 36/41 (86%)
Frame = +3
Query: 901 FVNAITEEATGFGPRPVFVIPGGIARDWDAIDIPSIMHVSD 941
FVNAI+EEATG G RP FV PGGIA DWDAIDIPSIMHVS+
Sbjct: 3 FVNAISEEATGSGLRPAFVTPGGIASDWDAIDIPSIMHVSE 125
Score = 32.3 bits (72), Expect = 1.2
Identities = 13/17 (76%), Positives = 14/17 (81%)
Frame = +3
Query: 951 FPVYEAEDEEGDDIPYE 967
FPVYE + EGDDIPYE
Sbjct: 483 FPVYEPKMREGDDIPYE 533
>BE124481
Length = 538
Score = 71.6 bits (174), Expect = 2e-12
Identities = 44/107 (41%), Positives = 52/107 (48%), Gaps = 11/107 (10%)
Frame = +3
Query: 267 PPFYHQYPLPPGQPQVPVNAIAQQMKQQLPVQQQQQNQQARPTFP-----------PIPM 315
PP Q P P QP P Q+ +QQ P Q QNQ + P PIP+
Sbjct: 231 PPLVQQRPQSPYQPMCP-----QKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPV 395
Query: 316 LYAELLPTLLHRGHCTTRQGKPPPDPLPPRFRSDLKCDFHQGALGHD 362
YA+LLP LL + T P+ LPP +R DL C FHQGA GHD
Sbjct: 396 KYADLLPILLKKNLIQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHD 536
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 48.5 bits (114), Expect = 2e-05
Identities = 22/49 (44%), Positives = 30/49 (60%)
Frame = -1
Query: 1091 ANIVPVPKKDGKVRMCVDFRDLNKASLKDNFPLPHIDVLVDNTAQSKVF 1139
A ++ V KKDG RMC+D+R NK + K+ +PLP ID L D + F
Sbjct: 238 AALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF 92
>TC92405 similar to GP|11994325|dbj|BAB02284. gene_id:MLM24.13~unknown
protein {Arabidopsis thaliana}, partial (69%)
Length = 864
Score = 38.1 bits (87), Expect = 0.023
Identities = 44/156 (28%), Positives = 64/156 (40%), Gaps = 7/156 (4%)
Frame = +2
Query: 658 SIVGNITACSNLWFSEDELP------EAGKHHNLALHISVNCKSDMISNVLVDTGSSLNV 711
SI G+I SN + S L +G + LA K+D + L D G +++
Sbjct: 152 SISGSIPIPSNPFDSTSPLKFNLLSAVSGLNRGLAASEEDLQKADAAAKELEDAGGLVDL 331
Query: 712 MPKSTLDQLSYRGTPLRRSTFLVKAFDGTRKSV-LGEIDLPITIGPETFLITFQVMDINA 770
LD+L R + S F + G+R +G + LPIT+G I D +
Sbjct: 332 T--DNLDRLQGRWKLIYSSAFSSRTLGGSRPGPPIGRL-LPITLGQVFQRIDILSKDFDN 502
Query: 771 SYSCLLGRPWIHDAGAVTSTLHQKLKFAKSGKLVTI 806
LG PW VT+TL K + S K+ I
Sbjct: 503 IVDLQLGAPWPLPPLEVTATLAHKFELVGSSKIKII 610
>BG648523 similar to GP|9955540|emb| putative protein {Arabidopsis thaliana},
partial (9%)
Length = 770
Score = 37.7 bits (86), Expect = 0.030
Identities = 26/73 (35%), Positives = 32/73 (43%), Gaps = 2/73 (2%)
Frame = +3
Query: 274 PLPPGQPQVPVNAIAQQMKQQLPVQQQQQNQQARPTFPPIPMLYAELLPTLLHRGHCT-- 331
PLPPG +N QQ QQ P Q Q QQ + P+PM +P + H H +
Sbjct: 411 PLPPGPHPSLLNPNQQQPFQQNPQQIPQHQQQLQQHMGPLPM--PPNMPQIQHSSHSSML 584
Query: 332 TRQGKPPPDPLPP 344
Q P P P P
Sbjct: 585 PHQHLPRPPPQMP 623
Score = 30.0 bits (66), Expect = 6.2
Identities = 26/125 (20%), Positives = 47/125 (36%), Gaps = 8/125 (6%)
Frame = +3
Query: 223 KKKETEIGMVSAGAGQPMATVALINAAQMPPSYPYVPYSQHPFFPPFYHQY--------P 274
++K+ G + A P +L+N Q P + Q+P P + Q P
Sbjct: 372 EQKQPHPGSIGAPPLPPGPHPSLLNPNQQQP------FQQNPQQIPQHQQQLQQHMGPLP 533
Query: 275 LPPGQPQVPVNAIAQQMKQQLPVQQQQQNQQARPTFPPIPMLYAELLPTLLHRGHCTTRQ 334
+PP PQ+ ++ + + Q + Q P P+P + +P + +
Sbjct: 534 MPPNMPQIQHSSHSSMLPHQHLPRPPPQMPHGMPGSLPVPTSHPMPIPGPMGMQGTMNQM 713
Query: 335 GKPPP 339
G P P
Sbjct: 714 GPPMP 728
>BQ144193 weakly similar to GP|14026007|dbj putative transporter
{Mesorhizobium loti}, partial (4%)
Length = 1037
Score = 36.6 bits (83), Expect = 0.066
Identities = 22/67 (32%), Positives = 29/67 (42%), Gaps = 1/67 (1%)
Frame = +2
Query: 214 SKRYGNGHHKK-KETEIGMVSAGAGQPMATVALINAAQMPPSYPYVPYSQHPFFPPFYHQ 272
+K+ NG K ET G + + P L+ +MPP YP HP PP H
Sbjct: 47 TKKRANGKQAKINETNAGKLKVKSNTP---AYLVGYKEMPPVYPRKSPKTHPRPPPNSHN 217
Query: 273 YPLPPGQ 279
P PP +
Sbjct: 218 RPEPPAK 238
>TC83704 similar to PIR|T05005|T05005 hypothetical protein T19P19.70 -
Arabidopsis thaliana, partial (15%)
Length = 716
Score = 35.0 bits (79), Expect = 0.19
Identities = 31/111 (27%), Positives = 45/111 (39%), Gaps = 6/111 (5%)
Frame = +1
Query: 241 ATVALINAAQMPPSYPYVPYSQHPFFPPFYHQYPLPPGQPQVPVNAIAQQMKQQLPVQQQ 300
+TV + +P PY QH PP P PP ++P A +Q+ P+ +
Sbjct: 262 STVPPVATTSLPEPLPY----QHKEQPPIPVTLPPPPPLSKLPP-ATREQLPPPPPLSKL 426
Query: 301 QQNQQARPTFPP----IPMLYAELLPTL--LHRGHCTTRQGKPPPDPLPPR 345
+ P PP +P E LP L + R+ PPP PLP +
Sbjct: 427 PPAAREHPPPPPPSAKLPPAARERLPLPPPLSKLPPAARERLPPPSPLPEK 579
>TC83149 similar to GP|13561982|gb|AAK30594.1 flagelliform silk protein
{Argiope trifasciata}, partial (3%)
Length = 1177
Score = 33.9 bits (76), Expect = 0.43
Identities = 29/107 (27%), Positives = 40/107 (37%), Gaps = 3/107 (2%)
Frame = +3
Query: 484 PPVKSMPLKENAEPLVIRLPGPVPYTSDRAIPYKYNATIIENGVEVPLVSLAVMNNTAEG 543
PP+ P P +R P P +S A P T P S +
Sbjct: 180 PPLPQHPKPPQKSPPRMRRSKPSP-SSPAATPLWPQRTTPPPSPSTPRPSP*TPATPSTS 356
Query: 544 TSAALRSGRVR---PPLFQKKAATPTVPPIDKPTPTDVSPVNRDASQ 587
+A L + R PP + +P+ PP KP P VSP + A+Q
Sbjct: 357 PTAPLPTPPPRTTLPPALTPRPPSPSTPPTPKPGPVSVSPASPSATQ 497
>TC91935
Length = 1144
Score = 33.9 bits (76), Expect = 0.43
Identities = 26/99 (26%), Positives = 45/99 (45%), Gaps = 8/99 (8%)
Frame = +2
Query: 30 VSKVDVPKKFKIPDFDRYNGLTCP--------QNHIIKYVRKMGNYKDNDSLMIHCFQDS 81
VS+V + ++ +P+F R LTCP N++I Y+ ++ N ++ C QD
Sbjct: 521 VSRVIIQSRYNVPNFSRSTALTCPTFDQEASLPNNLIAYLSRLANM----MIIFSCKQDH 688
Query: 82 LMEDAAEWYTSLSKNDIHTFDELAVAFKSHYGFNTRLKP 120
+ Y L I T ++ FK + +TR +P
Sbjct: 689 I-------YVRLKNKGIITLAQVE-GFKRN---DTRFQP 772
>TC77172 similar to GP|20466472|gb|AAM20553.1 putative protein {Arabidopsis
thaliana}, partial (78%)
Length = 4130
Score = 33.5 bits (75), Expect = 0.56
Identities = 25/90 (27%), Positives = 29/90 (31%), Gaps = 16/90 (17%)
Frame = +1
Query: 256 PYVPYSQHPFFPPFYHQYPLPPG----------------QPQVPVNAIAQQMKQQLPVQQ 299
P+ P Q F P P PP +PQ P + QQ +
Sbjct: 2680 PHQPPQQPNIFVPSQTTQPQPPQSNFPNTSGAQPPVRVFEPQTPALIRNPEQYQQPTLGS 2859
Query: 300 QQQNQQARPTFPPIPMLYAELLPTLLHRGH 329
Q N PTFPP Y P H GH
Sbjct: 2860 QLYNTNNNPTFPPTNQPYQPTPPAPSHIGH 2949
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,781,274
Number of Sequences: 36976
Number of extensions: 587258
Number of successful extensions: 3938
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 3584
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3871
length of query: 1323
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1215
effective length of database: 5,021,319
effective search space: 6100902585
effective search space used: 6100902585
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146794.16


