

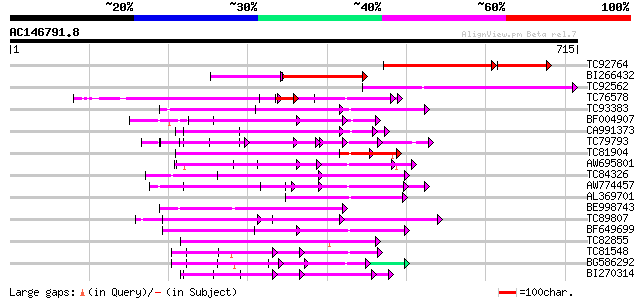
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146791.8 + phase: 0
(715 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC92764 similar to GP|8885599|dbj|BAA97529.1 contains similarity... 290 e-113
BI266432 similar to GP|8885599|dbj contains similarity to salt-i... 198 7e-51
TC92562 weakly similar to GP|9755888|emb|CAC01941.1 RSP67.2 {Rap... 163 2e-40
TC76578 weakly similar to GP|9755842|emb|CAC01928.1 67kD chlorop... 121 3e-30
TC93383 similar to PIR|T47786|T47786 hypothetical protein F17J16... 92 9e-19
BF004907 weakly similar to GP|8953393|emb putative protein {Arab... 92 9e-19
CA991373 weakly similar to PIR|D86260|D86 protein T12C24.22 [imp... 91 1e-18
TC79793 weakly similar to GP|6630464|gb|AAF19552.1| F23N19.4 {Ar... 91 2e-18
TC81904 weakly similar to GP|8778410|gb|AAF79418.1| F16A14.3 {Ar... 91 2e-18
AW695801 weakly similar to PIR|T01622|T01 probable salt-inducibl... 90 3e-18
TC84326 weakly similar to PIR|F85225|F85225 hypothetical protein... 87 2e-17
AW774457 weakly similar to GP|10176973|dbj gb|AAF19552.1~gene_id... 81 1e-15
AL369701 weakly similar to PIR|E84433|E8 hypothetical protein At... 80 4e-15
BE998743 similar to GP|22128591|gb| fertility restorer-like prot... 79 5e-15
TC89807 weakly similar to PIR|B96600|B96600 protein F14J16.14 [i... 79 5e-15
BF649699 similar to GP|9757911|dbj gb|AAF19552.1~gene_id:MMN10.1... 79 8e-15
TC82855 weakly similar to PIR|T45829|T45829 hypothetical protein... 75 9e-14
TC81548 weakly similar to GP|1931651|gb|AAB65486.1| membrane-ass... 75 1e-13
BG586292 similar to PIR|T46163|T46 nodulin / glutamate-ammonia l... 75 1e-13
BI270314 similar to GP|9759030|dbj contains similarity to salt-i... 73 3e-13
>TC92764 similar to GP|8885599|dbj|BAA97529.1 contains similarity to
salt-inducible protein~gene_id:F10E10.5 {Arabidopsis
thaliana}, partial (28%)
Length = 638
Score = 290 bits (742), Expect(2) = e-113
Identities = 142/143 (99%), Positives = 143/143 (99%)
Frame = +2
Query: 472 MGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLAC 531
MGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLAC
Sbjct: 2 MGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVLAC 181
Query: 532 LQRANPKLVAFIQLIVDEETSFETVKEEFKAIMSNAVVEVRRPFCNCLIDICRNKDLVER 591
LQRANPKLVAFIQLIVDEETSFETVKEEFKAIMSNAVVEVRRPFCNCLIDICRNKDLV+R
Sbjct: 182 LQRANPKLVAFIQLIVDEETSFETVKEEFKAIMSNAVVEVRRPFCNCLIDICRNKDLVQR 361
Query: 592 AHELLYLGTLYGFYPSLHNKTQY 614
AHELLYLGTLYGFYPSLHNKTQY
Sbjct: 362 AHELLYLGTLYGFYPSLHNKTQY 430
Score = 137 bits (346), Expect(2) = e-113
Identities = 67/68 (98%), Positives = 68/68 (99%)
Frame = +3
Query: 616 WCLDVRTLSVGAALTALEEWMTTLTKIVKREEALPDLFLAQTGTGAHKFAQGLNISFASH 675
WC+DVRTLSVGAALTALEEWMTTLTKIVKREEALPDLFLAQTGTGAHKFAQGLNISFASH
Sbjct: 435 WCIDVRTLSVGAALTALEEWMTTLTKIVKREEALPDLFLAQTGTGAHKFAQGLNISFASH 614
Query: 676 LRKLAAPF 683
LRKLAAPF
Sbjct: 615 LRKLAAPF 638
>BI266432 similar to GP|8885599|dbj contains similarity to salt-inducible
protein~gene_id:F10E10.5 {Arabidopsis thaliana}, partial
(14%)
Length = 345
Score = 198 bits (503), Expect = 7e-51
Identities = 95/108 (87%), Positives = 97/108 (88%)
Frame = +1
Query: 344 GKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILY 403
G G + + SGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILY
Sbjct: 4 GSRGLRGVCLRK*LTSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILY 183
Query: 404 NTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGA 451
NTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGA
Sbjct: 184 NTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGA 327
Score = 43.9 bits (102), Expect = 2e-04
Identities = 24/95 (25%), Positives = 48/95 (50%), Gaps = 1/95 (1%)
Frame = +1
Query: 254 TGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDG 313
+G+ P+E T +A++ +Y + ++ + L++R + GW D I ++ L M + G +
Sbjct: 49 SGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEE 228
Query: 314 IRYVLQEMK-SLGVQPNLVVYNTLLEAMGKAGKPG 347
+ ++MK S +P+ Y +L G G G
Sbjct: 229 AETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGACG 333
Score = 34.7 bits (78), Expect = 0.13
Identities = 15/15 (100%), Positives = 15/15 (100%)
Frame = +3
Query: 344 GKPGFARSLFEEMID 358
GKPGFARSLFEEMID
Sbjct: 3 GKPGFARSLFEEMID 47
>TC92562 weakly similar to GP|9755888|emb|CAC01941.1 RSP67.2 {Raphanus
sativus}, partial (17%)
Length = 1049
Score = 163 bits (412), Expect = 2e-40
Identities = 87/273 (31%), Positives = 154/273 (55%), Gaps = 3/273 (1%)
Frame = +2
Query: 446 YGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPD 505
Y G + +A + +EM G+E N++ T + C GKA +D+V +F+ ++ G+ PD
Sbjct: 2 YSWNGKIPEAEAMLKEMISCGLEPNILVLTMFVHCYGKAKRTNDVVNIFNQFMDTGITPD 181
Query: 506 DRLCGCLLSVVSLSQGSKDQEKVLACLQRANPKLVAFIQLIVDEETSFE--TVKEEFKAI 563
DRLC CLL V++ ++ K+ C+Q+ANP+L + ++ E + +E +
Sbjct: 182 DRLCDCLLYVMTQIP-KEEYGKITNCIQKANPRLGYIVTYLMKENEGDDDGNFMKETSEL 358
Query: 564 MSNAVVEVRRPFCNCLIDICRNKDLVERAHELLYLGTLYGFYPSLHNKTQYEWCLDVRTL 623
S+A +V++ CN LID+C L ++A LL LG + Y + ++++ +WCL+++ L
Sbjct: 359 *SSADDDVKKSLCNSLIDLCVKFGLQQKARNLLDLGLMLEIYSDIQSRSETQWCLNLKKL 538
Query: 624 SVGAALTALEEWMTTLTKIVKREEALPDLFLAQTGTGAHKFA-QGLNISFASHLRKLAAP 682
SVGAA+TA W+ L+K + E LP + T HK + + L F S+L++L +P
Sbjct: 539 SVGAAMTAFHVWIDDLSKAFESGEELPQVLGISTIPWRHKRSDKDLASVFESYLKELNSP 718
Query: 683 FRQSEDKVGCFIATKEDLISWVQSNSPSASIAT 715
F ++ + G F T ++ SW+QS + ++A+
Sbjct: 719 FHKATNMSGWFFTTSQEAKSWLQSRGSTETVAS 817
Score = 40.4 bits (93), Expect = 0.002
Identities = 16/58 (27%), Positives = 35/58 (59%)
Frame = +2
Query: 317 VLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKI 374
+L+EM S G++PN++V + GKA + ++F + +D+GI P+++ ++ +
Sbjct: 38 MLKEMISCGLEPNILVLTMFVHCYGKAKRTNDVVNIFNQFMDTGITPDDRLCDCLLYV 211
Score = 40.0 bits (92), Expect = 0.003
Identities = 21/70 (30%), Positives = 36/70 (51%)
Frame = +2
Query: 344 GKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILY 403
GK A ++ +EMI G+ PN LT + YGKA+ + D + ++ + + G D L
Sbjct: 14 GKIPEAEAMLKEMISCGLEPNILVLTMFVHCYGKAKRTNDVVNIFNQFMDTGITPDDRLC 193
Query: 404 NTLLNMCADV 413
+ LL + +
Sbjct: 194 DCLLYVMTQI 223
Score = 30.8 bits (68), Expect = 1.9
Identities = 17/68 (25%), Positives = 31/68 (45%)
Frame = +2
Query: 210 EELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDV 269
E + +MI G+E + + + + C K + V F + TG+ PD+ +L V
Sbjct: 32 EAMLKEMISCGLEPNILVLTMFVHCYGKAKRTNDVVNIFNQFMDTGITPDDRLCDCLLYV 211
Query: 270 YARLGKVE 277
++ K E
Sbjct: 212 MTQIPKEE 235
>TC76578 weakly similar to GP|9755842|emb|CAC01928.1 67kD chloroplastic
RNA-binding protein P67 {Arabidopsis thaliana}, partial
(27%)
Length = 1549
Score = 121 bits (303), Expect(2) = 3e-30
Identities = 83/263 (31%), Positives = 128/263 (48%)
Frame = +2
Query: 81 PENQPKVLSKPKPTWVNPTKTKRPVLSHQRHKRSSVSYNPQLREFQRFAQRLNNCDVSSS 140
P + LSK + WVNP KT +P KR + + + + LN VS
Sbjct: 743 PNSNSSSLSKSR-IWVNP-KTPKP-------KRIWNKSDKTVSSLAKLPKSLNEQQVSE- 892
Query: 141 DEEFMVCLEEIPSSLTRGNALLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSL 200
L + ++T +A +L+++ + + K + + YNV K
Sbjct: 893 ------ILNGLGDNVTERDAENILHNIADPETAIFVLEFFKQKIEFERHVVLYNVLFKLF 1054
Query: 201 RFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDE 260
R + F E+L +M+ GV+ D +T+ST+I CA C+ KAV FERM G PD
Sbjct: 1055REIKDFEQAEKLFDEMLQRGVKPDVVTFSTLIRCAAVCSFPHKAVELFERMPDFGCEPDY 1234
Query: 261 VTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQE 320
S+++ VYAR G V+ + L++ + W P FS L KM+G G+YDG V +
Sbjct: 1235NVSSSMIYVYARTGNVDMALKLYDSAKNEKWVIRPGAFSALIKMYGILGNYDGCLSVYND 1414
Query: 321 MKSLGVQPNLVVYNTLLEAMGKA 343
MK LGV+PN+V+YN + G++
Sbjct: 1415MKVLGVRPNMVLYNAFVGCYGES 1483
Score = 64.7 bits (156), Expect = 1e-10
Identities = 40/172 (23%), Positives = 89/172 (51%), Gaps = 1/172 (0%)
Frame = +2
Query: 316 YVLQEMKS-LGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKI 374
+VL+ K + + ++V+YN L + + A LF+EM+ G+ P+ T + +I+
Sbjct: 977 FVLEFFKQKIEFERHVVLYNVLFKLFREIKDFEQAEKLFDEMLQRGVKPDVVTFSTLIRC 1156
Query: 375 YGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKP 434
+ A+EL++RM + G D+ + ++++ + A G ++ A L+ D ++E
Sbjct: 1157 AAVCSFPHKAVELFERMPDFGCEPDYNVSSSMIYVYARTGNVDMALKLY-DSAKNEKWVI 1333
Query: 435 DSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAME 486
+++A++ +YG G D + ++ +M G+ N++ + C G++ E
Sbjct: 1334 RPGAFSALIKMYGILGNYDGCLSVYNDMKVLGVRPNMVLYNAFVGCYGES*E 1489
Score = 46.2 bits (108), Expect = 4e-05
Identities = 31/111 (27%), Positives = 55/111 (48%)
Frame = +2
Query: 385 LELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLN 444
LE +K+ E + +LYN L + ++ E+AE LF +M Q KPD +++ ++
Sbjct: 983 LEFFKQKIE--FERHVVLYNVLFKLFREIKDFEQAEKLFDEMLQ-RGVKPDVVTFSTLIR 1153
Query: 445 IYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFD 495
KA++LFE M FG E + + +I + +D +K++D
Sbjct: 1154 CAAVCSFPHKAVELFERMPDFGCEPDYNVSSSMIYVYARTGNVDMALKLYD 1306
Score = 33.1 bits (74), Expect = 0.38
Identities = 18/77 (23%), Positives = 39/77 (50%)
Frame = +2
Query: 439 YTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISV 498
Y + ++ ++A KLF+EM + G++ +V+ + LI+C V++F+
Sbjct: 1031 YNVLFKLFREIKDFEQAEKLFDEMLQRGVKPDVVTFSTLIRCAAVCSFPHKAVELFERMP 1210
Query: 499 ERGVKPDDRLCGCLLSV 515
+ G +PD + ++ V
Sbjct: 1211 DFGCEPDYNVSSSMIYV 1261
Score = 29.6 bits (65), Expect(2) = 3e-30
Identities = 12/29 (41%), Positives = 20/29 (68%)
Frame = +3
Query: 336 LLEAMGKAGKPGFARSLFEEMIDSGIAPN 364
LL+AMG+A + A+ +++EM +G PN
Sbjct: 1461 LLDAMGRAKRARDAKGVYQEMXKNGFVPN 1547
>TC93383 similar to PIR|T47786|T47786 hypothetical protein F17J16.90 -
Arabidopsis thaliana, partial (43%)
Length = 1038
Score = 91.7 bits (226), Expect = 9e-19
Identities = 55/220 (25%), Positives = 109/220 (49%)
Frame = +1
Query: 310 DYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLT 369
+Y + + +M+ ++P++V Y L+ A GKA + A ++FEEM+D+G+ P K
Sbjct: 40 NYKEVSNIYDQMQRADLRPDVVSYALLINAYGKARREEEALAVFEEMLDAGVRPTRKAYN 219
Query: 370 AVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQS 429
++ + + + A ++K M+ + + D Y T+L+ + +E AE F+ + Q
Sbjct: 220 ILLDAFSISGMVEQARIVFKSMRRDKYMPDLCSYTTMLSAYVNAPDMEGAEKFFKRLIQ- 396
Query: 430 EHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDD 489
+ +P+ +Y ++ Y ++K M+ +EEM GI+ N T ++ GK + D
Sbjct: 397 DGFEPNVVTYGTLIKGYAKANDIEKVMEKYEEMLGRGIKANQTILTTIMDAHGKNGDFDS 576
Query: 490 LVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVL 529
V F G+ PD + LLS+ + K+ +++
Sbjct: 577 AVNWFKEMALNGLLPDQKAKNILLSLAKTEEDIKEANELV 696
Score = 90.9 bits (224), Expect = 2e-18
Identities = 61/233 (26%), Positives = 107/233 (45%)
Frame = +1
Query: 190 TIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFE 249
T+ YN M F + + + QM + D ++Y+ +I+ K ++A+ FE
Sbjct: 4 TVTYNSLMS---FETNYKEVSNIYDQMQRADLRPDVVSYALLINAYGKARREEEALAVFE 174
Query: 250 RMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAG 309
M G+ P ++ +LD ++ G VE+ +F+ R + PD +++ + + A
Sbjct: 175 EMLDAGVRPTRKAYNILLDAFSISGMVEQARIVFKSMRRDKYMPDLCSYTTMLSAYVNAP 354
Query: 310 DYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLT 369
D +G + + G +PN+V Y TL++ KA +EEM+ GI N+ LT
Sbjct: 355 DMEGAEKFFKRLIQDGFEPNVVTYGTLIKGYAKANDIEKVMEKYEEMLGRGIKANQTILT 534
Query: 370 AVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETL 422
++ +GK A+ +K M NG D N LL++ I+EA L
Sbjct: 535 TIMDAHGKNGDFDSAVNWFKEMALNGLLPDQKAKNILLSLAKTEEDIKEANEL 693
>BF004907 weakly similar to GP|8953393|emb putative protein {Arabidopsis
thaliana}, partial (34%)
Length = 665
Score = 91.7 bits (226), Expect = 9e-19
Identities = 55/201 (27%), Positives = 97/201 (47%)
Frame = +3
Query: 226 ITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFER 285
+TY T++ + +KA+ M K G+ P+ + ++ I+D A G+ +E + + ER
Sbjct: 30 VTYGTLVEGYCRMRRVEKALEMVGEMTKEGIKPNAIVYNPIIDALAEAGRFKEALGMMER 209
Query: 286 GRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGK 345
P T++ L K F +AGD +G +L++M S G P YN + GK
Sbjct: 210 FHVLQIGPTLSTYNSLVKGFCKAGDIEGASKILKKMISRGFLPIPTTYNYFFRYFSRCGK 389
Query: 346 PGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNT 405
+L+ +MI+SG P+ T V+K+ + + A+++ M+ G+ MD
Sbjct: 390 VDEGMNLYTKMIESGHNPDRLTYHLVLKMLCEEEKLELAVQVSMEMRHKGYDMDLATSTM 569
Query: 406 LLNMCADVGLIEEAETLFRDM 426
L ++ + +EEA F DM
Sbjct: 570 LTHLLCKMHKLEEAFAEFEDM 632
Score = 73.6 bits (179), Expect = 3e-13
Identities = 48/210 (22%), Positives = 98/210 (45%)
Frame = +3
Query: 258 PDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYV 317
P VT+ +++ Y R+ +VE+ + + G KP+ I ++ + EAG + +
Sbjct: 21 PSVVTYGTLVEGYCRMRRVEKALEMVGEMTKEGIKPNAIVYNPIIDALAEAGRFKEALGM 200
Query: 318 LQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGK 377
++ L + P L YN+L++ KAG A + ++MI G P T + + +
Sbjct: 201 MERFHVLQIGPTLSTYNSLVKGFCKAGDIEGASKILKKMISRGFLPIPTTYNYFFRYFSR 380
Query: 378 ARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSW 437
+ + L+ +M E+G D + Y+ +L M + +E A + +M+ + D
Sbjct: 381 CGKVDEGMNLYTKMIESGHNPDRLTYHLVLKMLCEEEKLELAVQVSMEMRHKGY-DMDLA 557
Query: 438 SYTAMLNIYGSEGAVDKAMKLFEEMSKFGI 467
+ T + ++ +++A FE+M + GI
Sbjct: 558 TSTMLTHLLCKMHKLEEAFAEFEDMIRRGI 647
Score = 61.2 bits (147), Expect = 1e-09
Identities = 56/220 (25%), Positives = 96/220 (43%), Gaps = 4/220 (1%)
Frame = +3
Query: 152 PSSLTRGNALLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSL----RFGRQFG 207
PS +T G + +R +K + + + P I YN + +L RF G
Sbjct: 21 PSVVTYGTLVEGYCRMRRVEKALEMVGEMTKEGIKP-NAIVYNPIIDALAEAGRFKEALG 197
Query: 208 IIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAIL 267
++E ++ G L TY++++ K + A ++M G +P T++
Sbjct: 198 MMERF--HVLQIGPTLS--TYNSLVKGFCKAGDIEGASKILKKMISRGFLPIPTTYNYFF 365
Query: 268 DVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQ 327
++R GKV+E +NL+ + +G PD +T+ ++ KM E + V EM+ G
Sbjct: 366 RYFSRCGKVDEGMNLYTKMIESGHNPDRLTYHLVLKMLCEEEKLELAVQVSMEMRHKGYD 545
Query: 328 PNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKT 367
+L L + K K A + FE+MI GI P T
Sbjct: 546 MDLATSTMLTHLLCKMHKLEEAFAEFEDMIRRGIIPQYLT 665
Score = 39.3 bits (90), Expect = 0.005
Identities = 28/148 (18%), Positives = 66/148 (43%)
Frame = +3
Query: 358 DSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIE 417
+ + P+ T +++ Y + R + ALE+ M + G + I+YN +++ A+ G +
Sbjct: 6 NENVRPSVVTYGTLVEGYCRMRRVEKALEMVGEMTKEGIKPNAIVYNPIIDALAEAGRFK 185
Query: 418 EAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCL 477
EA + + P +Y +++ + G ++ A K+ ++M G
Sbjct: 186 EALGMMERFHVLQ-IGPTLSTYNSLVKGFCKAGDIEGASKILKKMISRGFLPIPTTYNYF 362
Query: 478 IQCLGKAMEIDDLVKVFDISVERGVKPD 505
+ + ++D+ + ++ +E G PD
Sbjct: 363 FRYFSRCGKVDEGMNLYTKMIESGHNPD 446
>CA991373 weakly similar to PIR|D86260|D86 protein T12C24.22 [imported] -
Arabidopsis thaliana, partial (6%)
Length = 756
Score = 91.3 bits (225), Expect = 1e-18
Identities = 63/245 (25%), Positives = 123/245 (49%)
Frame = +2
Query: 220 GVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEV 279
G++ +TY +++ +KA M + G+ PD +++ ++D + ++ KV+E
Sbjct: 11 GIKPIVVTYCSLMDGYCLVKEVNKAKSILYTMSQRGVNPDIQSYNILIDGFCKIKKVDEA 190
Query: 280 VNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEA 339
+NLF+ PD +T++ L + G ++ EM GV P+++ Y+++L+A
Sbjct: 191 MNLFKEMHHKHIIPDVVTYNSLIDGLCKLGKISYALKLVDEMHDRGVPPDIITYSSILDA 370
Query: 340 MGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMD 399
+ K + A +L ++ D GI PN T T +I K +DA +++ + G+ +
Sbjct: 371 LCKNHQVDKAIALLTKLKDQGIRPNMYTYTILIDGLCKGGRLEDAHNIFEDLLVKGYNIT 550
Query: 400 FILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLF 459
Y +++ + GL +EA TL MK + C PD+ +Y ++ + DKA KL
Sbjct: 551 VNTYTVMIHGFCNKGLFDEALTLLSKMKDNS-CIPDAVTYEIIIRSLFDKDENDKAEKLR 727
Query: 460 EEMSK 464
E +++
Sbjct: 728 EMITR 742
Score = 89.7 bits (221), Expect = 3e-18
Identities = 55/213 (25%), Positives = 104/213 (48%)
Frame = +2
Query: 210 EELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDV 269
+ + + M GV D +Y+ +I K D+A+ F+ M+ ++PD VT+++++D
Sbjct: 86 KSILYTMSQRGVNPDIQSYNILIDGFCKIKKVDEAMNLFKEMHHKHIIPDVVTYNSLIDG 265
Query: 270 YARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPN 329
+LGK+ + L + G PD IT+S + + D +L ++K G++PN
Sbjct: 266 LCKLGKISYALKLVDEMHDRGVPPDIITYSSILDALCKNHQVDKAIALLTKLKDQGIRPN 445
Query: 330 LVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWK 389
+ Y L++ + K G+ A ++FE+++ G T T +I + +AL L
Sbjct: 446 MYTYTILIDGLCKGGRLEDAHNIFEDLLVKGYNITVNTYTVMIHGFCNKGLFDEALTLLS 625
Query: 390 RMKENGWPMDFILYNTLLNMCADVGLIEEAETL 422
+MK+N D + Y ++ D ++AE L
Sbjct: 626 KMKDNSCIPDAVTYEIIIRSLFDKDENDKAEKL 724
Score = 68.2 bits (165), Expect = 1e-11
Identities = 49/189 (25%), Positives = 90/189 (46%)
Frame = +2
Query: 290 GWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFA 349
G KP +T+ L + + + + +L M GV P++ YN L++ K K A
Sbjct: 11 GIKPIVVTYCSLMDGYCLVKEVNKAKSILYTMSQRGVNPDIQSYNILIDGFCKIKKVDEA 190
Query: 350 RSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNM 409
+LF+EM I P+ T ++I K AL+L M + G P D I Y+++L+
Sbjct: 191 MNLFKEMHHKHIIPDVVTYNSLIDGLCKLGKISYALKLVDEMHDRGVPPDIITYSSILDA 370
Query: 410 CADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIEL 469
+++A L +K + +P+ ++YT +++ G ++ A +FE++ G +
Sbjct: 371 LCKNHQVDKAIALLTKLKD-QGIRPNMYTYTILIDGLCKGGRLEDAHNIFEDLLVKGYNI 547
Query: 470 NVMGCTCLI 478
V T +I
Sbjct: 548 TVNTYTVMI 574
Score = 30.0 bits (66), Expect = 3.2
Identities = 25/116 (21%), Positives = 44/116 (37%)
Frame = +2
Query: 152 PSSLTRGNALLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEE 211
P +T + L L K +K Q + P Y + + L G +
Sbjct: 335 PDIITYSSILDALCKNHQVDKAIALLTKLKDQGIRP-NMYTYTILIDGLCKGGRLEDAHN 511
Query: 212 LAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAIL 267
+ ++ G + TY+ +I LFD+A+ +M +PD VT+ I+
Sbjct: 512 IFEDLLVKGYNITVNTYTVMIHGFCNKGLFDEALTLLSKMKDNSCIPDAVTYEIII 679
>TC79793 weakly similar to GP|6630464|gb|AAF19552.1| F23N19.4 {Arabidopsis
thaliana}, partial (7%)
Length = 1580
Score = 90.9 bits (224), Expect = 2e-18
Identities = 60/236 (25%), Positives = 116/236 (48%)
Frame = +1
Query: 191 IFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFER 250
+ YN M ++ + + + M GGV D +YS +I+ K FD+A+ F+
Sbjct: 559 VTYNSLMDGYCLVKEVNKAKSIFNTMAQGGVNPDIQSYSIMINGFCKIKKFDEAMNLFKE 738
Query: 251 MYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGD 310
M++ ++PD VT+S+++D ++ G++ + L ++ G P+ T++ + +
Sbjct: 739 MHRKNIIPDVVTYSSLIDGLSKSGRISYALQLVDQMHDRGVPPNICTYNSILDALCKTHQ 918
Query: 311 YDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTA 370
D +L + K G QP++ Y+ L++ + ++GK AR +FE ++ G N T T
Sbjct: 919 VDKAIALLTKFKDKGFQPDISTYSILIKGLCQSGKLEDARKVFEGLLVKGHNLNVDTYTI 1098
Query: 371 VIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDM 426
+I+ + +AL L +M++NG D Y ++ + AE L R+M
Sbjct: 1099MIQGFCVEGLFNEALALLSKMEDNGCIPDAKTYEIIILSLFKKDENDMAEKLLREM 1266
Score = 87.8 bits (216), Expect = 1e-17
Identities = 62/251 (24%), Positives = 121/251 (47%)
Frame = +1
Query: 220 GVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEV 279
G++ + +TY++++ +KA F M + G+ PD ++S +++ + ++ K +E
Sbjct: 541 GIKPNFVTYNSLMDGYCLVKEVNKAKSIFNTMAQGGVNPDIQSYSIMINGFCKIKKFDEA 720
Query: 280 VNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEA 339
+NLF+ PD +T+S L ++G ++ +M GV PN+ YN++L+A
Sbjct: 721 MNLFKEMHRKNIIPDVVTYSSLIDGLSKSGRISYALQLVDQMHDRGVPPNICTYNSILDA 900
Query: 340 MGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMD 399
+ K + A +L + D G P+ T + +IK ++ +DA ++++ + G ++
Sbjct: 901 LCKTHQVDKAIALLTKFKDKGFQPDISTYSILIKGLCQSGKLEDARKVFEGLLVKGHNLN 1080
Query: 400 FILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLF 459
Y ++ GL EA L M + C PD+ +Y ++ + D A KL
Sbjct: 1081VDTYTIMIQGFCVEGLFNEALALLSKM-EDNGCIPDAKTYEIIILSLFKKDENDMAEKLL 1257
Query: 460 EEMSKFGIELN 470
EM G+ N
Sbjct: 1258REMIARGLP*N 1290
Score = 70.9 bits (172), Expect = 2e-12
Identities = 51/208 (24%), Positives = 97/208 (46%)
Frame = +1
Query: 190 TIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFE 249
+I N K +F + +E+ + I V +TYS++I K A+ +
Sbjct: 673 SIMINGFCKIKKFDEAMNLFKEMHRKNIIPDV----VTYSSLIDGLSKSGRISYALQLVD 840
Query: 250 RMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAG 309
+M+ G+ P+ T+++ILD + +V++ + L + + G++PD T+S+L K ++G
Sbjct: 841 QMHDRGVPPNICTYNSILDALCKTHQVDKAIALLTKFKDKGFQPDISTYSILIKGLCQSG 1020
Query: 310 DYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLT 369
+ R V + + G N+ Y +++ G A +L +M D+G P+ KT
Sbjct: 1021KLEDARKVFEGLLVKGHNLNVDTYTIMIQGFCVEGLFNEALALLSKMEDNGCIPDAKTYE 1200
Query: 370 AVIKIYGKARWSKDALELWKRMKENGWP 397
+I K + A +L + M G P
Sbjct: 1201IIILSLFKKDENDMAEKLLREMIARGLP 1284
Score = 69.3 bits (168), Expect = 5e-12
Identities = 41/139 (29%), Positives = 74/139 (52%)
Frame = +2
Query: 253 KTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYD 312
K G +PD +TF+ + G++++ ++ A G+ D I++ L + G+
Sbjct: 8 KMGYVPDTITFTTLSKGLCLKGQIQQAFLFHDKVVALGFHFDQISYGTLIHGLCKVGETR 187
Query: 313 GIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVI 372
+LQ + VQPN+V+YNT++++M K A LF EM+ GI+P+ T +A+I
Sbjct: 188 AALDLLQRVDGNLVQPNVVMYNTIIDSMCKVKLVNEAFDLFSEMVSKGISPDVVTYSALI 367
Query: 373 KIYGKARWSKDALELWKRM 391
+ KDA++L+ +M
Sbjct: 368 SGFCILGKLKDAIDLFNKM 424
Score = 68.2 bits (165), Expect = 1e-11
Identities = 58/245 (23%), Positives = 109/245 (43%)
Frame = +1
Query: 290 GWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFA 349
G KP+ +T++ L + + + + + M GV P++ Y+ ++ K K A
Sbjct: 541 GIKPNFVTYNSLMDGYCLVKEVNKAKSIFNTMAQGGVNPDIQSYSIMINGFCKIKKFDEA 720
Query: 350 RSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNM 409
+LF+EM I P+ T +++I K+ AL+L +M + G P + YN++L+
Sbjct: 721 MNLFKEMHRKNIIPDVVTYSSLIDGLSKSGRISYALQLVDQMHDRGVPPNICTYNSILDA 900
Query: 410 CADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIEL 469
+++A L K +PD +Y+ ++ G ++ A K+FE + G L
Sbjct: 901 LCKTHQVDKAIALLTKFKDKGF-QPDISTYSILIKGLCQSGKLEDARKVFEGLLVKGHNL 1077
Query: 470 NVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKVL 529
NV T +IQ ++ + + + G PD + ++ LS KD+ +
Sbjct: 1078NVDTYTIMIQGFCVEGLFNEALALLSKMEDNGCIPDAK----TYEIIILSLFKKDENDMA 1245
Query: 530 ACLQR 534
L R
Sbjct: 1246EKLLR 1260
Score = 65.1 bits (157), Expect = 9e-11
Identities = 45/198 (22%), Positives = 91/198 (45%)
Frame = +1
Query: 167 LRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNI 226
++ + + F + +N++P + + Y+ + L + +L QM D GV +
Sbjct: 700 IKKFDEAMNLFKEMHRKNIIP-DVVTYSSLIDGLSKSGRISYALQLVDQMHDRGVPPNIC 876
Query: 227 TYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERG 286
TY++I+ K + DKA+ + G PD T+S ++ + GK+E+ +FE
Sbjct: 877 TYNSILDALCKTHQVDKAIALLTKFKDKGFQPDISTYSILIKGLCQSGKLEDARKVFEGL 1056
Query: 287 RATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKP 346
G + T++++ + F G ++ +L +M+ G P+ Y ++ ++ K +
Sbjct: 1057LVKGHNLNVDTYTIMIQGFCVEGLFNEALALLSKMEDNGCIPDAKTYEIIILSLFKKDEN 1236
Query: 347 GFARSLFEEMIDSGIAPN 364
A L EMI G+ N
Sbjct: 1237DMAEKLLREMIARGLP*N 1290
Score = 57.0 bits (136), Expect = 2e-08
Identities = 31/89 (34%), Positives = 49/89 (54%), Gaps = 1/89 (1%)
Frame = +2
Query: 215 QMIDGGVELDNIT-YSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARL 273
Q +DG + N+ Y+TII K L ++A F M G+ PD VT+SA++ + L
Sbjct: 206 QRVDGNLVQPNVVMYNTIIDSMCKVKLVNEAFDLFSEMVSKGISPDVVTYSALISGFCIL 385
Query: 274 GKVEEVVNLFERGRATGWKPDPITFSVLG 302
GK+++ ++LF + KPD TF +G
Sbjct: 386 GKLKDAIDLFNKMILENIKPDVYTF*YIG 472
>TC81904 weakly similar to GP|8778410|gb|AAF79418.1| F16A14.3 {Arabidopsis
thaliana}, partial (7%)
Length = 978
Score = 90.5 bits (223), Expect = 2e-18
Identities = 66/251 (26%), Positives = 117/251 (46%)
Frame = +1
Query: 210 EELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDV 269
E + +M G+ D Y ++ FDKA+ ++ M G+ + V FS IL
Sbjct: 22 ESVFLEMEKQGLVPDVYVYCALVHGYCNSRNFDKALAVYKSMISRGIKTNCVIFSCILHC 201
Query: 270 YARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPN 329
+G+ EVV++FE + +G D +++L + G D +L E+KS+ + +
Sbjct: 202 LDEMGRALEVVDMFEEFKESGLFIDRKAYNILFDALCKLGKVDDAVGMLDELKSMQLDVD 381
Query: 330 LVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWK 389
+ Y TL+ GKP A+SLF+EM + G P+ + + + R +A++L
Sbjct: 382 MKHYTTLINGYFLQGKPIEAQSLFKEMEERGFKPDVVAYNVLAAGFFRNRTDFEAMDLLN 561
Query: 390 RMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSE 449
M+ G + + ++ G +EEAE F +K E + YTA++N Y
Sbjct: 562 YMESQGVEPNSTTHKIIIEGLCSAGKVEEAEEFFNWLK-GESVEISVEIYTALVNGYCEA 738
Query: 450 GAVDKAMKLFE 460
++K+ +L E
Sbjct: 739 ALIEKSHELKE 771
Score = 48.5 bits (114), Expect = 9e-06
Identities = 25/82 (30%), Positives = 52/82 (62%), Gaps = 3/82 (3%)
Frame = +1
Query: 416 IEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCT 475
++EAE++F +M++ + PD + Y A+++ Y + DKA+ +++ M GI+ N + +
Sbjct: 10 LDEAESVFLEMEK-QGLVPDVYVYCALVHGYCNSRNFDKALAVYKSMISRGIKTNCVIFS 186
Query: 476 CLIQCL---GKAMEIDDLVKVF 494
C++ CL G+A+E+ D+ + F
Sbjct: 187 CILHCLDEMGRALEVVDMFEEF 252
Score = 32.3 bits (72), Expect = 0.65
Identities = 31/149 (20%), Positives = 54/149 (35%)
Frame = +1
Query: 152 PSSLTRGNALLVLNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEE 211
P+S T + L S ++ FFNW+K G I E
Sbjct: 586 PNSTTHKIIIEGLCSAGKVEEAEEFFNWLK---------------------GESVEISVE 702
Query: 212 LAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYA 271
+ +++G E + +K + +A M + + P +V +S I
Sbjct: 703 IYTALVNGYCE---------AALIEKSHELKEAFILLRTMLEMNMKPSKVMYSKIFTALC 855
Query: 272 RLGKVEEVVNLFERGRATGWKPDPITFSV 300
G +E LF TG+ PD +T+++
Sbjct: 856 CNGNMEGAHTLFNLFXHTGFTPDAVTYTI 942
Score = 30.4 bits (67), Expect = 2.5
Identities = 17/65 (26%), Positives = 31/65 (47%)
Frame = +1
Query: 449 EGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRL 508
E +D+A +F EM K G+ +V L+ + D + V+ + RG+K + +
Sbjct: 1 ETKLDEAESVFLEMEKQGLVPDVYVYCALVHGYCNSRNFDKALAVYKSMISRGIKTNCVI 180
Query: 509 CGCLL 513
C+L
Sbjct: 181 FSCIL 195
>AW695801 weakly similar to PIR|T01622|T01 probable salt-inducible protein
At2g18940 [imported] - Arabidopsis thaliana, partial
(18%)
Length = 655
Score = 90.1 bits (222), Expect = 3e-18
Identities = 52/185 (28%), Positives = 99/185 (53%)
Frame = +3
Query: 209 IEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILD 268
+E HQ+ + G +LD + ++++S + +KA + ++ +GL P+ VT+++++D
Sbjct: 15 MERAFHQLQNNGYKLDMVVINSMLSMFVRNQKLEKAHEMLDVIHVSGLQPNLVTYNSLID 194
Query: 269 VYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQP 328
+YAR+G + + + + +G PD ++++ + K F + G +L EM + GVQP
Sbjct: 195 LYARVGDCWKAEEMLKDIQNSGISPDVVSYNTVIKGFCKKGLVQEAIRILSEMTANGVQP 374
Query: 329 NLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELW 388
+ +NT + G A + MI+ G PNE T VI Y KA+ K+A++
Sbjct: 375 CPITFNTFMSCYAGNGLFAEADEVIRYMIEHGCMPNELTYKIVIDGYIKAKKHKEAMDFV 554
Query: 389 KRMKE 393
++KE
Sbjct: 555 SKIKE 569
Score = 73.9 bits (180), Expect = 2e-13
Identities = 51/206 (24%), Positives = 98/206 (46%), Gaps = 1/206 (0%)
Frame = +3
Query: 283 FERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGK 342
F + + G+K D + + + MF + +L + G+QPNLV YN+L++ +
Sbjct: 27 FHQLQNNGYKLDMVVINSMLSMFVRNQKLEKAHEMLDVIHVSGLQPNLVTYNSLIDLYAR 206
Query: 343 AGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFIL 402
G A + +++ +SGI+P+ + VIK + K ++A+ + M NG I
Sbjct: 207 VGDCWKAEEMLKDIQNSGISPDVVSYNTVIKGFCKKGLVQEAIRILSEMTANGVQPCPIT 386
Query: 403 YNTLLNMCADVGLIEEAETLFRDMKQSEH-CKPDSWSYTAMLNIYGSEGAVDKAMKLFEE 461
+NT ++ A GL EA+ + R M EH C P+ +Y +++ Y +AM +
Sbjct: 387 FNTFMSCYAGNGLFAEADEVIRYM--IEHGCMPNELTYKIVIDGYIKAKKHKEAMDFVSK 560
Query: 462 MSKFGIELNVMGCTCLIQCLGKAMEI 487
+ + I + L C+ +++ +
Sbjct: 561 IKEIDISFDDQSLKKLASCIRESLGV 638
Score = 61.2 bits (147), Expect = 1e-09
Identities = 38/162 (23%), Positives = 80/162 (48%), Gaps = 4/162 (2%)
Frame = +3
Query: 211 ELAHQMID----GGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAI 266
E AH+M+D G++ + +TY+++I + KA + + +G+ PD V+++ +
Sbjct: 114 EKAHEMLDVIHVSGLQPNLVTYNSLIDLYARVGDCWKAEEMLKDIQNSGISPDVVSYNTV 293
Query: 267 LDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGV 326
+ + + G V+E + + A G +P PITF+ + G + V++ M G
Sbjct: 294 IKGFCKKGLVQEAIRILSEMTANGVQPCPITFNTFMSCYAGNGLFAEADEVIRYMIEHGC 473
Query: 327 QPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTL 368
PN + Y +++ KA K A ++ + I+ ++++L
Sbjct: 474 MPNELTYKIVIDGYIKAKKHKEAMDFVSKIKEIDISFDDQSL 599
Score = 55.5 bits (132), Expect = 7e-08
Identities = 43/204 (21%), Positives = 94/204 (46%), Gaps = 4/204 (1%)
Frame = +3
Query: 313 GIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVI 372
G+ ++++ G + ++VV N++L + K A + + + SG+ PN T ++I
Sbjct: 12 GMERAFHQLQNNGYKLDMVVINSMLSMFVRNQKLEKAHEMLDVIHVSGLQPNLVTYNSLI 191
Query: 373 KIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHC 432
+Y + A E+ K ++ +G D + YNT++ GL++EA + +M +
Sbjct: 192 DLYARVGDCWKAEEMLKDIQNSGISPDVVSYNTVIKGFCKKGLVQEAIRILSEM-TANGV 368
Query: 433 KPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAME----ID 488
+P ++ ++ Y G +A ++ M + G N + +I KA + +D
Sbjct: 369 QPCPITFNTFMSCYAGNGLFAEADEVIRYMIEHGCMPNELTYKIVIDGYIKAKKHKEAMD 548
Query: 489 DLVKVFDISVERGVKPDDRLCGCL 512
+ K+ +I + + +L C+
Sbjct: 549 FVSKIKEIDISFDDQSLKKLASCI 620
>TC84326 weakly similar to PIR|F85225|F85225 hypothetical protein AT4g19900
[imported] - Arabidopsis thaliana, partial (18%)
Length = 756
Score = 87.4 bits (215), Expect = 2e-17
Identities = 61/242 (25%), Positives = 115/242 (47%)
Frame = +1
Query: 263 FSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMK 322
++A++ Y R K+ L R + G P+ T++ L +AG+++ ++ M
Sbjct: 1 YTAMIRGYCREDKLNRAEMLLSRMKEQGLVPNTNTYTTLIDGHCKAGNFERAYDLMNLMS 180
Query: 323 SLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSK 382
S G PNL YN ++ + K G+ A + E+ +G+ P++ T ++ + K +
Sbjct: 181 SEGFSPNLCTYNAIVNGLCKRGRVQEAYKMLEDGFQNGLKPDKFTYNILMSEHCKQENIR 360
Query: 383 DALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAM 442
AL L+ +M + G D Y TL+ + ++E+E F + + P + +YT+M
Sbjct: 361 QALALFNKMLKIGIQPDIHSYTTLIAVFCRENRMKESEMFFEEAVRI-GIIPTNKTYTSM 537
Query: 443 LNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGV 502
+ Y EG + AMK F +S G + +I L K + D+ ++D +E+G+
Sbjct: 538 ICGYCREGNLTLAMKFFHRLSDHGCAPESITYGAIISGLCKQSKRDEARSLYDSMIEKGL 717
Query: 503 KP 504
P
Sbjct: 718 VP 723
Score = 62.4 bits (150), Expect = 6e-10
Identities = 48/224 (21%), Positives = 91/224 (40%)
Frame = +1
Query: 172 KTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTI 231
+ M + +K Q L+P + + + G F +L + M G + TY+ I
Sbjct: 46 RAEMLLSRMKEQGLVPNTNTYTTLIDGHCKAGN-FERAYDLMNLMSSEGFSPNLCTYNAI 222
Query: 232 ISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGW 291
++ K +A E ++ GL PD+ T++ ++ + + + + + LF + G
Sbjct: 223 VNGLCKRGRVQEAYKMLEDGFQNGLKPDKFTYNILMSEHCKQENIRQALALFNKMLKIGI 402
Query: 292 KPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARS 351
+PD +++ L +F +E +G+ P Y +++ + G A
Sbjct: 403 QPDIHSYTTLIAVFCRENRMKESEMFFEEAVRIGIIPTNKTYTSMICGYCREGNLTLAMK 582
Query: 352 LFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENG 395
F + D G AP T A+I K +A L+ M E G
Sbjct: 583 FFHRLSDHGCAPESITYGAIISGLCKQSKRDEARSLYDSMIEKG 714
Score = 36.6 bits (83), Expect = 0.034
Identities = 25/92 (27%), Positives = 45/92 (48%)
Frame = +1
Query: 171 QKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYST 230
+++ MFF ++P + ++ R G + + H++ D G ++ITY
Sbjct: 463 KESEMFFEEAVRIGIIPTNKTYTSMICGYCREGN-LTLAMKFFHRLSDHGCAPESITYGA 639
Query: 231 IISCAKKCNLFDKAVYWFERMYKTGLMPDEVT 262
IIS K + D+A ++ M + GL+P EVT
Sbjct: 640 IISGLCKQSKRDEARSLYDSMIEKGLVPCEVT 735
>AW774457 weakly similar to GP|10176973|dbj
gb|AAF19552.1~gene_id:MTG13.9~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (13%)
Length = 650
Score = 81.3 bits (199), Expect = 1e-15
Identities = 53/186 (28%), Positives = 93/186 (49%)
Frame = -1
Query: 317 VLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYG 376
+ +M+ G+ P+ V YN+L++ + K+G+ +A L +EM D+G N T +I
Sbjct: 602 LFNDMQFKGIAPDKVTYNSLIDGLCKSGRISYAWELVDEMHDNGQPANIFTYNCLIDALC 423
Query: 377 KARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDS 436
K A+ L K++K+ G D +N L+ VG ++ A+ +F+D+ S+ ++
Sbjct: 422 KNHHVDQAIALVKKIKDQGIQPDMYTFNILIYGLCKVGRLKNAQDVFQDL-LSKGYSVNA 246
Query: 437 WSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDI 496
W+Y M+N EG D+A L +M GI + + LIQ L E + K+
Sbjct: 245 WTYNIMVNGLCKEGLFDEAEALLSKMDDNGIIPDAVTYETLIQALFHKDENEKAEKLLRE 66
Query: 497 SVERGV 502
+ RG+
Sbjct: 65 MIARGL 48
Score = 68.9 bits (167), Expect = 6e-12
Identities = 43/185 (23%), Positives = 91/185 (48%)
Frame = -1
Query: 177 FNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAK 236
FN ++ + + P + + YN + L + EL +M D G + TY+ +I
Sbjct: 599 FNDMQFKGIAP-DKVTYNSLIDGLCKSGRISYAWELVDEMHDNGQPANIFTYNCLIDALC 423
Query: 237 KCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPI 296
K + D+A+ +++ G+ PD TF+ ++ ++G+++ ++F+ + G+ +
Sbjct: 422 KNHHVDQAIALVKKIKDQGIQPDMYTFNILIYGLCKVGRLKNAQDVFQDLLSKGYSVNAW 243
Query: 297 TFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEM 356
T++++ + G +D +L +M G+ P+ V Y TL++A+ + A L EM
Sbjct: 242 TYNIMVNGLCKEGLFDEAEALLSKMDDNGIIPDAVTYETLIQALFHKDENEKAEKLLREM 63
Query: 357 IDSGI 361
I G+
Sbjct: 62 IARGL 48
Score = 62.4 bits (150), Expect = 6e-10
Identities = 42/176 (23%), Positives = 81/176 (45%)
Frame = -1
Query: 220 GVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEV 279
G+ D +TY+++I K A + M+ G + T++ ++D + V++
Sbjct: 578 GIAPDKVTYNSLIDGLCKSGRISYAWELVDEMHDNGQPANIFTYNCLIDALCKNHHVDQA 399
Query: 280 VNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEA 339
+ L ++ + G +PD TF++L + G + V Q++ S G N YN ++
Sbjct: 398 IALVKKIKDQGIQPDMYTFNILIYGLCKVGRLKNAQDVFQDLLSKGYSVNAWTYNIMVNG 219
Query: 340 MGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENG 395
+ K G A +L +M D+GI P+ T +I+ ++ A +L + M G
Sbjct: 218 LCKEGLFDEAEALLSKMDDNGIIPDAVTYETLIQALFHKDENEKAEKLLREMIARG 51
Score = 55.8 bits (133), Expect = 5e-08
Identities = 42/181 (23%), Positives = 81/181 (44%)
Frame = -1
Query: 349 ARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLN 408
A SLF +M GIAP++ T ++I K+ A EL M +NG P + YN L++
Sbjct: 611 ALSLFNDMQFKGIAPDKVTYNSLIDGLCKSGRISYAWELVDEMHDNGQPANIFTYNCLID 432
Query: 409 MCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIE 468
+++A L + +K + +PD +++ ++ G + A +F+++ G
Sbjct: 431 ALCKNHHVDQAIALVKKIK-DQGIQPDMYTFNILIYGLCKVGRLKNAQDVFQDLLSKGYS 255
Query: 469 LNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGCLLSVVSLSQGSKDQEKV 528
+N ++ L K D+ + + G+ PD L+ + ++ EK+
Sbjct: 254 VNAWTYNIMVNGLCKEGLFDEAEALLSKMDDNGIIPDAVTYETLIQALFHKDENEKAEKL 75
Query: 529 L 529
L
Sbjct: 74 L 72
Score = 28.9 bits (63), Expect = 7.2
Identities = 19/65 (29%), Positives = 28/65 (42%)
Frame = -1
Query: 193 YNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMY 252
YN+ + L F E L +M D G+ D +TY T+I + +KA M
Sbjct: 239 YNIMVNGLCKEGLFDEAEALLSKMDDNGIIPDAVTYETLIQALFHKDENEKAEKLLREMI 60
Query: 253 KTGLM 257
GL+
Sbjct: 59 ARGLL 45
>AL369701 weakly similar to PIR|E84433|E8 hypothetical protein At2g02150
[imported] - Arabidopsis thaliana, partial (11%)
Length = 463
Score = 79.7 bits (195), Expect = 4e-15
Identities = 47/153 (30%), Positives = 80/153 (51%)
Frame = +1
Query: 349 ARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLN 408
A LF ++++G+A N + T++I Y KA+ + A+++ K M + + +D LY T +
Sbjct: 7 AEELFRALLEAGVALNLQIYTSLIHGYMKAKMMEKAMDILKEMNKKNFKLDSPLYGTKIW 186
Query: 409 MCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIE 468
IEE+E + R+MK +S+ YT++++ Y G + +A+ L +EM KFGIE
Sbjct: 187 GLCSQNKIEESEAVMREMKD-HGLTANSYIYTSLMDAYFKVGKITEAVNLLQEMQKFGIE 363
Query: 469 LNVMGCTCLIQCLGKAMEIDDLVKVFDISVERG 501
+ LI L K + V FD + G
Sbjct: 364 TTAVTYGVLIDGLCKKGLVQQAVSYFDCMTKTG 462
Score = 40.4 bits (93), Expect = 0.002
Identities = 33/150 (22%), Positives = 58/150 (38%)
Frame = +1
Query: 211 ELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVY 270
++ +M +LD+ Y T I N +++ M GL + +++++D Y
Sbjct: 118 DILKEMNKKNFKLDSPLYGTKIWGLCSQNKIEESEAVMREMKDHGLTANSYIYTSLMDAY 297
Query: 271 ARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNL 330
++GK+ E VNL LQEM+ G++
Sbjct: 298 FKVGKITEAVNL-----------------------------------LQEMQKFGIETTA 372
Query: 331 VVYNTLLEAMGKAGKPGFARSLFEEMIDSG 360
V Y L++ + K G A S F+ M +G
Sbjct: 373 VTYGVLIDGLCKKGLVQQAVSYFDCMTKTG 462
>BE998743 similar to GP|22128591|gb| fertility restorer-like protein {Petunia
x hybrida}, partial (7%)
Length = 829
Score = 79.3 bits (194), Expect = 5e-15
Identities = 56/237 (23%), Positives = 115/237 (47%)
Frame = +2
Query: 190 TIFYNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFE 249
TI ++ ++ + G G LA MI G ++ D +TY++++ N +KA +
Sbjct: 74 TINPSLPLRMRKEGNVKGAKNALA-MMIKGSIKPDVVTYNSLMDGYCLVNEVNKAKHVLS 250
Query: 250 RMYKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAG 309
+ + G+ PD +++ +++ + R+ ++V + G PD T+S L +
Sbjct: 251 TIARMGVAPDAQSYNIMVNGFCRISHAWKLV---DEMHVNGQPPDIFTYSSLIDALCKNN 421
Query: 310 DYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLT 369
D +++++K G+QPN+ YN L++ + K G+ A+ +F++++ G + N +T
Sbjct: 422 HLDKAIALVKKIKDQGIQPNMYTYNILIDGLCKGGRLKNAQDVFQDLLTKGYSLNIRTYN 601
Query: 370 AVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDM 426
+I K A L +M++N + + Y T++ E+AE L R+M
Sbjct: 602 ILINGLCKEGLFDKAEALLSKMEDNDINPNVVTYETIIRSLFYKDYNEKAEKLLREM 772
>TC89807 weakly similar to PIR|B96600|B96600 protein F14J16.14 [imported] -
Arabidopsis thaliana, partial (53%)
Length = 1241
Score = 79.3 bits (194), Expect = 5e-15
Identities = 55/232 (23%), Positives = 115/232 (48%), Gaps = 2/232 (0%)
Frame = +1
Query: 193 YNVTMKSLRFGRQFGIIEELA-HQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERM 251
Y T++ L ++F + ++ HQ + + + + +I+ K N+ A F+ M
Sbjct: 166 YEDTVRRLAGAKRFRWVRDIIEHQKSYADISNEGFS-ARLITLYGKSNMHRHAQKLFDEM 342
Query: 252 YKTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRAT-GWKPDPITFSVLGKMFGEAGD 310
+ ++ +A+L Y + + V LF++ KPD ++++ K E G
Sbjct: 343 PQRNCERSVLSLNALLAAYLHSKQYDVVERLFKKLPVQLSVKPDLVSYNTYIKALLEKGS 522
Query: 311 YDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTA 370
+D VL+EM+ GV+ +L+ +NTLL+ + G+ L+E++ + + PN +T A
Sbjct: 523 FDSAVSVLEEMEKDGVESDLITFNTLLDGLYSKGRFEDGEKLWEKLGEKNVVPNIRTYNA 702
Query: 371 VIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETL 422
+ A+ + +A+E ++ M++ G D +N L+ A+ G ++EA+ +
Sbjct: 703 RLLGLAVAKRAGEAVEFYEEMEKKGVKPDLFSFNALIKGFANEGNLDEAKNV 858
Score = 59.3 bits (142), Expect = 5e-09
Identities = 47/217 (21%), Positives = 92/217 (41%), Gaps = 2/217 (0%)
Frame = +1
Query: 332 VYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRM 391
+Y + + A + + R + E NE +I +YGK+ + A +L+ M
Sbjct: 163 IYEDTVRRLAGAKRFRWVRDIIEHQKSYADISNEGFSARLITLYGKSNMHRHAQKLFDEM 342
Query: 392 KENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYGSEGA 451
+ + N LL + E LF+ + KPD SY + +G+
Sbjct: 343 PQRNCERSVLSLNALLAAYLHSKQYDVVERLFKKLPVQLSVKPDLVSYNTYIKALLEKGS 522
Query: 452 VDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKPDDRLCGC 511
D A+ + EEM K G+E +++ L+ L +D K+++ E+ V P+ R
Sbjct: 523 FDSAVSVLEEMEKDGVESDLITFNTLLDGLYSKGRFEDGEKLWEKLGEKNVVPNIRTYNA 702
Query: 512 LLSVVSLSQGSKDQEKVLACLQR--ANPKLVAFIQLI 546
L +++++ + + + +++ P L +F LI
Sbjct: 703 RLLGLAVAKRAGEAVEFYEEMEKKGVKPDLFSFNALI 813
Score = 52.0 bits (123), Expect = 8e-07
Identities = 42/162 (25%), Positives = 74/162 (44%), Gaps = 2/162 (1%)
Frame = +1
Query: 159 NALLV--LNSLRPWQKTHMFFNWIKTQNLLPMETIFYNVTMKSLRFGRQFGIIEELAHQM 216
NALL L+S + + F + Q + + + YN +K+L F + +M
Sbjct: 379 NALLAAYLHS-KQYDVVERLFKKLPVQLSVKPDLVSYNTYIKALLEKGSFDSAVSVLEEM 555
Query: 217 IDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKV 276
GVE D IT++T++ F+ +E++ + ++P+ T++A L A +
Sbjct: 556 EKDGVESDLITFNTLLDGLYSKGRFEDGEKLWEKLGEKNVVPNIRTYNARLLGLAVAKRA 735
Query: 277 EEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVL 318
E V +E G KPD +F+ L K F G+ D + V+
Sbjct: 736 GEAVEFYEEMEKKGVKPDLFSFNALIKGFANEGNLDEAKNVV 861
>BF649699 similar to GP|9757911|dbj gb|AAF19552.1~gene_id:MMN10.14~similar to
unknown protein {Arabidopsis thaliana}, partial (11%)
Length = 617
Score = 78.6 bits (192), Expect = 8e-15
Identities = 58/196 (29%), Positives = 96/196 (48%)
Frame = +1
Query: 309 GDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTL 368
G+ D ++ ++ G PNL VYN L+ A+ K A L++ M + N+ T
Sbjct: 16 GNIDSAYDLVVKLGRFGFLPNLFVYNALINALCKGEDLDKAELLYKNMHSMNLPLNDVTY 195
Query: 369 TAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQ 428
+ +I + K A + RM E+G YN+L+N G + AE L+ M
Sbjct: 196 SILIDSFCKRGMLDVAESYFGRMIEDGIRETIYPYNSLINGHCKFGDLSAAEFLYTKMI- 372
Query: 429 SEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEID 488
+E +P + ++T +++ Y + V+KA KL+ EM++ +V T LI L E+
Sbjct: 373 NEGLEPTATTFTTLISGYCKDLQVEKAFKLYREMNEKEXAPSVYTFTALIYGLCSTNEMA 552
Query: 489 DLVKVFDISVERGVKP 504
+ K+FD VER +KP
Sbjct: 553 EASKLFDEMVERKIKP 600
Score = 55.5 bits (132), Expect = 7e-08
Identities = 38/175 (21%), Positives = 76/175 (42%)
Frame = +1
Query: 193 YNVTMKSLRFGRQFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMY 252
YN + +L G E L M + L+++TYS +I K + D A +F RM
Sbjct: 88 YNALINALCKGEDLDKAELLYKNMHSMNLPLNDVTYSILIDSFCKRGMLDVAESYFGRMI 267
Query: 253 KTGLMPDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYD 312
+ G+ ++++++ + + G + L+ + G +P TF+ L + + +
Sbjct: 268 EDGIRETIYPYNSLINGHCKFGDLSAAEFLYTKMINEGLEPTATTFTTLISGYCKDLQVE 447
Query: 313 GIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKT 367
+ +EM P++ + L+ + + A LF+EM++ I P T
Sbjct: 448 KAFKLYREMNEKEXAPSVYTFTALIYGLCSTNEMAEASKLFDEMVERKIKPTXVT 612
>TC82855 weakly similar to PIR|T45829|T45829 hypothetical protein F2K15.100
- Arabidopsis thaliana, partial (63%)
Length = 1447
Score = 75.1 bits (183), Expect = 9e-14
Identities = 62/260 (23%), Positives = 119/260 (44%), Gaps = 8/260 (3%)
Frame = +3
Query: 216 MIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTG-LMPDEVTFSAILDVYARLG 274
+ GV + ITY+ I C D A+ F++ K P TF ++
Sbjct: 33 LTQAGVVPNIITYNLIFQTYLDCRKPDTALENFKQFIKDAPFNPSPTTFRILIKGLVDNN 212
Query: 275 KVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSL--GVQPNLVV 332
+++ +++ ++ A+G+ PDP+ + L + D DG+ V +E+K GV + VV
Sbjct: 213 RLDRAMDIKDQMNASGFAPDPLVYHYLMLGHARSSDGDGVLRVYEELKEKLGGVVEDGVV 392
Query: 333 YNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRM- 391
L++ G A ++E+ G ++ +V+ K +A+ L+ RM
Sbjct: 393 LGCLMKGYFLKGMEKEAMECYQEVFVEGKKMSDIAYNSVLDALAKNGKFDEAMRLFDRMI 572
Query: 392 KENGWPMDFIL----YNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNIYG 447
KE+ P + +N +++ G +EA +FR M +S CKPD+ S+ ++
Sbjct: 573 KEHNPPAKLAVNLGSFNVMVDGYCAEGKFKEAIEVFRSMGES-RCKPDTLSFNNLIEQLC 749
Query: 448 SEGAVDKAMKLFEEMSKFGI 467
+ G + +A +++ EM G+
Sbjct: 750 NNGMILEAEEVYGEMEGKGV 809
Score = 34.7 bits (78), Expect = 0.13
Identities = 22/82 (26%), Positives = 43/82 (51%), Gaps = 1/82 (1%)
Frame = +1
Query: 319 QEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKA 378
++M G++PNL VYN L++ + K GK A+ F+ M+ + + + ++K+ +A
Sbjct: 892 KKMVESGLRPNLAVYNRLVDGLVKVGKIDDAKFFFDLMVKK-LKMDVASYQFMMKVLSEA 1068
Query: 379 RWSKDALELWKR-MKENGWPMD 399
D L++ + +NG D
Sbjct: 1069GRLDDVLQIVNMLLDDNGVDFD 1134
>TC81548 weakly similar to GP|1931651|gb|AAB65486.1| membrane-associated
salt-inducible protein isolog; 88078-84012 {Arabidopsis
thaliana}, partial (23%)
Length = 882
Score = 74.7 bits (182), Expect = 1e-13
Identities = 60/208 (28%), Positives = 97/208 (45%), Gaps = 1/208 (0%)
Frame = +1
Query: 264 SAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKS 323
+A L +A K V LF K D ++S K D + + +++
Sbjct: 241 NATLHHFANSNKFNHVSQLF-LWMQENKKLDVYSYSHYIKFMANKLDASTMLKLYNDIQV 417
Query: 324 LGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWS-K 382
+ N+ V N++L + K GK LF +M G+ P+ T + +I K +
Sbjct: 418 ESAKDNVYVCNSVLSCLIKKGKFDTTMKLFRQMKHDGLVPDLVTYSTLIAGCVKVKDGYP 597
Query: 383 DALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAM 442
ALEL + +++N MD ++Y +L +CA G EEAE F MK SE P+ + Y+++
Sbjct: 598 KALELIQELQDNKLRMDDVIYGAILAVCASNGKWEEAEYYFNQMK-SEGRSPNVYHYSSL 774
Query: 443 LNIYGSEGAVDKAMKLFEEMSKFGIELN 470
LN Y + G KA L ++M G+ N
Sbjct: 775 LNAYSACGDFTKADALIQDMESEGLAPN 858
Score = 65.9 bits (159), Expect = 5e-11
Identities = 43/134 (32%), Positives = 69/134 (51%), Gaps = 1/134 (0%)
Frame = +1
Query: 205 QFGIIEELAHQMIDGGVELDNITYSTIIS-CAKKCNLFDKAVYWFERMYKTGLMPDEVTF 263
+F +L QM G+ D +TYST+I+ C K + + KA+ + + L D+V +
Sbjct: 481 KFDTTMKLFRQMKHDGLVPDLVTYSTLIAGCVKVKDGYPKALELIQELQDNKLRMDDVIY 660
Query: 264 SAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKS 323
AIL V A GK EE F + ++ G P+ +S L + GD+ ++Q+M+S
Sbjct: 661 GAILAVCASNGKWEEAEYYFNQMKSEGRSPNVYHYSSLLNAYSACGDFTKADALIQDMES 840
Query: 324 LGVQPNLVVYNTLL 337
G+ PN + TLL
Sbjct: 841 EGLAPNKGILTTLL 882
Score = 65.5 bits (158), Expect = 7e-11
Identities = 43/154 (27%), Positives = 77/154 (49%), Gaps = 5/154 (3%)
Frame = +1
Query: 224 DNI-TYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEE---- 278
DN+ ++++SC K FD + F +M GL+PD VT+S ++ A KV++
Sbjct: 430 DNVYVCNSVLSCLIKKGKFDTTMKLFRQMKHDGLVPDLVTYSTLI---AGCVKVKDGYPK 600
Query: 279 VVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLE 338
+ L + + + D + + + + G ++ Y +MKS G PN+ Y++LL
Sbjct: 601 ALELIQELQDNKLRMDDVIYGAILAVCASNGKWEEAEYYFNQMKSEGRSPNVYHYSSLLN 780
Query: 339 AMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVI 372
A G A +L ++M G+APN+ LT ++
Sbjct: 781 AYSACGDFTKADALIQDMESEGLAPNKGILTTLL 882
Score = 41.6 bits (96), Expect = 0.001
Identities = 34/145 (23%), Positives = 64/145 (43%), Gaps = 1/145 (0%)
Frame = +1
Query: 264 SAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGD-YDGIRYVLQEMK 322
+++L + GK + + LF + + G PD +T+S L + D Y ++QE++
Sbjct: 448 NSVLSCLIKKGKFDTTMKLFRQMKHDGLVPDLVTYSTLIAGCVKVKDGYPKALELIQELQ 627
Query: 323 SLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKARWSK 382
++ + V+Y +L GK A F +M G +PN ++++ Y
Sbjct: 628 DNKLRMDDVIYGAILAVCASNGKWEEAEYYFNQMKSEGRSPNVYHYSSLLNAYSACGDFT 807
Query: 383 DALELWKRMKENGWPMDFILYNTLL 407
A L + M+ G + + TLL
Sbjct: 808 KADALIQDMESEGLAPNKGILTTLL 882
>BG586292 similar to PIR|T46163|T46 nodulin / glutamate-ammonia ligase-like
protein - Arabidopsis thaliana, partial (50%)
Length = 719
Score = 74.7 bits (182), Expect = 1e-13
Identities = 43/142 (30%), Positives = 74/142 (51%), Gaps = 1/142 (0%)
Frame = -3
Query: 205 QFGIIEELAHQMIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTG-LMPDEVTF 263
+F ++ + +M GV +TY+TI+ K LF++ M + G +PD T
Sbjct: 468 RFDLVNRVLLEMSYLGVRCSTVTYNTILDGYGKAGLFEEMENVLADMIEDGDSLPDVFTL 289
Query: 264 SAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKS 323
++I+ Y G + ++ + + R + G +PD TF+VL FG+AG Y + V+ M+
Sbjct: 288 NSIIGSYGNGGNIRKMESWYNRFQLMGVEPDITTFNVLILSFGKAGMYKKMSSVMDYMEK 109
Query: 324 LGVQPNLVVYNTLLEAMGKAGK 345
P +V YN ++E GKAG+
Sbjct: 108 RFFSPTVVTYNIVIETFGKAGR 43
Score = 71.2 bits (173), Expect = 1e-12
Identities = 49/194 (25%), Positives = 92/194 (47%), Gaps = 2/194 (1%)
Frame = -3
Query: 263 FSAILDVYARLGKVEEVVNLFERGR-ATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEM 321
+++++ VY + +E+ E + A+ PD TF+VL + +D + VL EM
Sbjct: 612 YTSLISVYGKCSLLEKAFATLEYMKSASDCTPDVFTFTVLISCCCKLARFDLVNRVLLEM 433
Query: 322 KSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIA-PNEKTLTAVIKIYGKARW 380
LGV+ + V YNT+L+ GKAG ++ +MI+ G + P+ TL ++I YG
Sbjct: 432 SYLGVRCSTVTYNTILDGYGKAGLFEEMENVLADMIEDGDSLPDVFTLNSIIGSYGNGGN 253
Query: 381 SKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYT 440
+ + R + G D +N L+ G+ ++ ++ D + P +Y
Sbjct: 252 IRKMESWYNRFQLMGVEPDITTFNVLILSFGKAGMYKKMSSVM-DYMEKRFFSPTVVTYN 76
Query: 441 AMLNIYGSEGAVDK 454
++ +G G ++K
Sbjct: 75 IVIETFGKAGRIEK 34
Score = 64.7 bits (156), Expect = 1e-10
Identities = 45/159 (28%), Positives = 73/159 (45%), Gaps = 4/159 (2%)
Frame = -3
Query: 224 DNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEVVN-- 281
D T++ +ISC K FD M G+ VT++ ILD Y + G EE+ N
Sbjct: 516 DVFTFTVLISCCCKLARFDLVNRVLLEMSYLGVRCSTVTYNTILDGYGKAGLFEEMENVL 337
Query: 282 --LFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEA 339
+ E G + PD T + + +G G+ + + +GV+P++ +N L+ +
Sbjct: 336 ADMIEDGDSL---PDVFTLNSIIGSYGNGGNIRKMESWYNRFQLMGVEPDITTFNVLILS 166
Query: 340 MGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYGKA 378
GKAG S+ + M +P T VI+ +GKA
Sbjct: 165 FGKAGMYKKMSSVMDYMEKRFFSPTVVTYNIVIETFGKA 49
Score = 56.6 bits (135), Expect = 3e-08
Identities = 45/179 (25%), Positives = 71/179 (39%), Gaps = 1/179 (0%)
Frame = -3
Query: 327 QPNLVVYNTLLEAMGKAGKPGFARSLFEEMID-SGIAPNEKTLTAVIKIYGKARWSKDAL 385
+P + VY +L+ GK A + E M S P+ T T +I K
Sbjct: 630 KPTIDVYTSLISVYGKCSLLEKAFATLEYMKSASDCTPDVFTFTVLISCCCKLARFDLVN 451
Query: 386 ELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDSWSYTAMLNI 445
+ M G + YNT+L+ GL EE E + DM + PD ++ +++
Sbjct: 450 RVLLEMSYLGVRCSTVTYNTILDGYGKAGLFEEMENVLADMIEDGDSLPDVFTLNSIIGS 271
Query: 446 YGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVKVFDISVERGVKP 504
YG+ G + K + G+E ++ LI GKA + V D +R P
Sbjct: 270 YGNGGNIRKMESWYNRFQLMGVEPDITTFNVLILSFGKAGMYKKMSSVMDYMEKRFFSP 94
>BI270314 similar to GP|9759030|dbj contains similarity to salt-inducible
protein~gene_id:K6A12.14 {Arabidopsis thaliana}, partial
(30%)
Length = 669
Score = 73.2 bits (178), Expect = 3e-13
Identities = 53/195 (27%), Positives = 86/195 (43%), Gaps = 3/195 (1%)
Frame = +3
Query: 292 KPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEAMGKAGK-PGFAR 350
KP +TF++L + + +L EMK G++PN Y L+ A G+ K A
Sbjct: 3 KPTAVTFNILMYAYSRRMQPKIVESLLAEMKDFGLKPNANSYTCLISAYGRQKKMSDMAA 182
Query: 351 SLFEEMIDSGIAPNEKTLTAVIKIYGKARWSKDALELWKRMKENGWPMDFILYNTLLNMC 410
F +M GI P + TA+I Y + W + A +++ M G Y TLL+
Sbjct: 183 DAFLKMKKVGIKPTSHSYTAMIHAYSVSGWHEKAYAVFENMIREGIKPSIETYTTLLDAF 362
Query: 411 ADVGLIEEAETLFRDMK--QSEHCKPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIE 468
VG + ETL + K SE K ++ +++ + +G +A + E K G++
Sbjct: 363 RRVG---DTETLMKIWKLMMSEKVKGTQVTFNILVDGFAKQGLFMEARDVISEFGKIGLQ 533
Query: 469 LNVMGCTCLIQCLGK 483
VM LI +
Sbjct: 534 PTVMTYNMLINAYAR 578
Score = 71.2 bits (173), Expect = 1e-12
Identities = 40/153 (26%), Positives = 76/153 (49%)
Frame = +3
Query: 220 GVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGKVEEV 279
G++ + +Y+ +I +KA FE M + G+ P T++ +LD + R+G E +
Sbjct: 210 GIKPTSHSYTAMIHAYSVSGWHEKAYAVFENMIREGIKPSIETYTTLLDAFRRVGDTETL 389
Query: 280 VNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNTLLEA 339
+ +++ + K +TF++L F + G + R V+ E +G+QP ++ YN L+ A
Sbjct: 390 MKIWKLMMSEKVKGTQVTFNILVDGFAKQGLFMEARDVISEFGKIGLQPTVMTYNMLINA 569
Query: 340 MGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVI 372
+ G L +EM + P+ T + VI
Sbjct: 570 YARGGLDSNIPQLLKEMEALXLRPDSITYSTVI 668
Score = 68.9 bits (167), Expect = 6e-12
Identities = 49/206 (23%), Positives = 95/206 (45%), Gaps = 1/206 (0%)
Frame = +3
Query: 258 PDEVTFSAILDVYARLGKVEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDY-DGIRY 316
P VTF+ ++ Y+R + + V +L + G KP+ +++ L +G D
Sbjct: 6 PTAVTFNILMYAYSRRMQPKIVESLLAEMKDFGLKPNANSYTCLISAYGRQKKMSDMAAD 185
Query: 317 VLQEMKSLGVQPNLVVYNTLLEAMGKAGKPGFARSLFEEMIDSGIAPNEKTLTAVIKIYG 376
+MK +G++P Y ++ A +G A ++FE MI GI P+ +T T ++ +
Sbjct: 186 AFLKMKKVGIKPTSHSYTAMIHAYSVSGWHEKAYAVFENMIREGIKPSIETYTTLLDAFR 365
Query: 377 KARWSKDALELWKRMKENGWPMDFILYNTLLNMCADVGLIEEAETLFRDMKQSEHCKPDS 436
+ ++ +++WK M + +N L++ A GL EA + + + +P
Sbjct: 366 RVGDTETLMKIWKLMMSEKVKGTQVTFNILVDGFAKQGLFMEARDVISEFGKI-GLQPTV 542
Query: 437 WSYTAMLNIYGSEGAVDKAMKLFEEM 462
+Y ++N Y G +L +EM
Sbjct: 543 MTYNMLINAYARGGLDSNIPQLLKEM 620
Score = 48.9 bits (115), Expect = 7e-06
Identities = 27/122 (22%), Positives = 64/122 (52%)
Frame = +3
Query: 216 MIDGGVELDNITYSTIISCAKKCNLFDKAVYWFERMYKTGLMPDEVTFSAILDVYARLGK 275
MI G++ TY+T++ ++ + + ++ M + +VTF+ ++D +A+ G
Sbjct: 303 MIREGIKPSIETYTTLLDAFRRVGDTETLMKIWKLMMSEKVKGTQVTFNILVDGFAKQGL 482
Query: 276 VEEVVNLFERGRATGWKPDPITFSVLGKMFGEAGDYDGIRYVLQEMKSLGVQPNLVVYNT 335
E ++ G +P +T+++L + G I +L+EM++L ++P+ + Y+T
Sbjct: 483 FMEARDVISEFGKIGLQPTVMTYNMLINAYARGGLDSNIPQLLKEMEALXLRPDSITYST 662
Query: 336 LL 337
++
Sbjct: 663 VI 668
Score = 38.5 bits (88), Expect = 0.009
Identities = 38/181 (20%), Positives = 74/181 (39%), Gaps = 5/181 (2%)
Frame = +3
Query: 433 KPDSWSYTAMLNIYGSEGAVDKAMKLFEEMSKFGIELNVMGCTCLIQCLGKAMEIDDLVK 492
KP + ++ ++ Y L EM FG++ N TCLI G+ ++ D+
Sbjct: 3 KPTAVTFNILMYAYSRRMQPKIVESLLAEMKDFGLKPNANSYTCLISAYGRQKKMSDMAA 182
Query: 493 VFDISVER-GVKPDDRLCGCLLSVVSLSQGSKDQEKVLACLQR--ANPKLVAFIQLI--V 547
+ +++ G+KP ++ S+S + V + R P + + L+
Sbjct: 183 DAFLKMKKVGIKPTSHSYTAMIHAYSVSGWHEKAYAVFENMIREGIKPSIETYTTLLDAF 362
Query: 548 DEETSFETVKEEFKAIMSNAVVEVRRPFCNCLIDICRNKDLVERAHELLYLGTLYGFYPS 607
ET+ + +K +MS V + F N L+D + L A +++ G P+
Sbjct: 363 RRVGDTETLMKIWKLMMSEKVKGTQVTF-NILVDGFAKQGLFMEARDVISEFGKIGLQPT 539
Query: 608 L 608
+
Sbjct: 540 V 542
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,537,047
Number of Sequences: 36976
Number of extensions: 328374
Number of successful extensions: 2430
Number of sequences better than 10.0: 163
Number of HSP's better than 10.0 without gapping: 2115
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2333
length of query: 715
length of database: 9,014,727
effective HSP length: 103
effective length of query: 612
effective length of database: 5,206,199
effective search space: 3186193788
effective search space used: 3186193788
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC146791.8


