

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146790.6 - phase: 0
(360 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
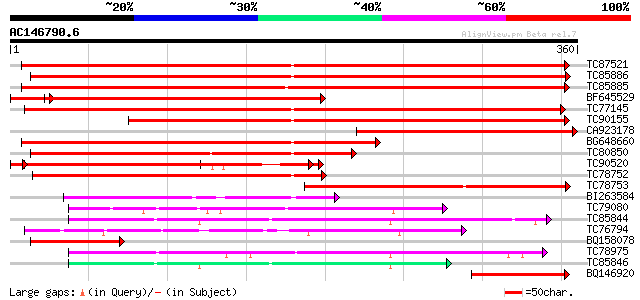
Score E
Sequences producing significant alignments: (bits) Value
TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 433 e-122
TC85886 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 423 e-119
TC85885 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 422 e-119
BF645529 weakly similar to GP|22475166|gb putative sinapyl alcoh... 372 e-113
TC77145 homologue to SP|P31656|CADH_MEDSA Cinnamyl-alcohol dehyd... 350 4e-97
TC90155 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydro... 309 9e-85
CA923178 weakly similar to SP|P93257|MTD_ Probable mannitol dehy... 276 9e-75
BG648660 similar to SP|Q9ZRF1|MTD_ Probable mannitol dehydrogena... 276 9e-75
TC80850 homologue to SP|O82515|MTD_MEDSA Probable mannitol dehyd... 272 1e-73
TC90520 weakly similar to PIR|T08581|T08581 cinnamyl-alcohol deh... 246 2e-67
TC78752 similar to GP|22857590|gb|AAN09864.1 putative cinnamyl-a... 209 1e-54
TC78753 similar to PIR|E96751|E96751 hypothetical protein F28P22... 177 7e-45
BI263584 similar to SP|Q9P6C8|ADH1 Alcohol dehydrogenase I (EC 1... 122 2e-28
TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogena... 92 4e-19
TC85844 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 ... 90 2e-18
TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbi... 84 8e-17
BQ158078 similar to GP|10187159|emb cDNA~Strawberry alcohol dehy... 83 1e-16
TC78975 similar to GP|21592656|gb|AAM64605.1 alcohol dehydrogena... 82 3e-16
TC85846 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 ... 73 2e-13
BQ146920 similar to GP|8099340|gb|A ELI3 {Lycopersicon esculentu... 72 3e-13
>TC87521 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (98%)
Length = 1433
Score = 433 bits (1113), Expect = e-122
Identities = 207/348 (59%), Positives = 262/348 (74%)
Frame = +2
Query: 8 HTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPV 67
H GWAA D+SG ++P+ F RRE G DV K+LYCGICH+D+H K++WG++ YP+
Sbjct: 92 HPNKAFGWAARDTSGVLSPFNFSRRETGEKDVAFKVLYCGICHSDLHMIKNEWGMSTYPL 271
Query: 68 VPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIF 127
VPGHEI G++T+VGS VE FK GDKVGVGCL SC C+ C+ + ENYC K Y+ +
Sbjct: 272 VPGHEIAGIVTEVGSKVEKFKIGDKVGVGCLVDSCRACQNCEENLENYCPKQTNTYSAKY 451
Query: 128 WDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRI 187
DGSITYGGYS +V D ++VHIP+ LP+++AAPLLCAGITV+SPL+ GL G I
Sbjct: 452 SDGSITYGGYSDSMVADEHFIVHIPDGLPLESAAPLLCAGITVYSPLRYFGL-DKPGMNI 628
Query: 188 GVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRS 247
G+VGLGGLGH+ VKF KAFG +VTVISTSP+K+ EA E LGAD F+IS + D++QAA +
Sbjct: 629 GIVGLGGLGHLGVKFAKAFGANVTVISTSPNKEKEAIENLGADSFLISHDQDKMQAAMGT 808
Query: 248 LDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIK 307
LD I+DTVSADH LLP++ LLK +G L +VGAPDKP +LP PLI G+++I G IGG+K
Sbjct: 809 LDGIIDTVSADHPLLPLVGLLKYHGKLVMVGAPDKPPELPHIPLIMGRKTISGSGIGGMK 988
Query: 308 ETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIAN 355
ETQEM+D KHNI DIE+I D +N A +RLLK DV+YRFV+DI N
Sbjct: 989 ETQEMIDFAAKHNIKPDIEVIPVDYVNTAMKRLLKADVKYRFVLDIGN 1132
>TC85886 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (94%)
Length = 1395
Score = 423 bits (1087), Expect = e-119
Identities = 203/343 (59%), Positives = 258/343 (75%)
Frame = +2
Query: 14 GWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEI 73
GWAA DSSG ++P+ F RRE G DV K+LYCGICH+D+H +++WG T YP+VPGHE+
Sbjct: 113 GWAARDSSGVLSPFHFFRRETGEKDVAFKVLYCGICHSDLHIMQNEWGKTTYPLVPGHEL 292
Query: 74 TGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSIT 133
TGV+T+VGS V+ FK GDKVGVG + SC CE C D ENYC K +NG DG+IT
Sbjct: 293 TGVVTEVGSKVKKFKVGDKVGVGYMVDSCRSCENCADDIENYCTKYTQTFNGKSRDGTIT 472
Query: 134 YGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLG 193
YGG+S +V D +V+ IP+SLP+D A PLLCAG+TV+SPL+ GL G IGVVGLG
Sbjct: 473 YGGFSDSMVADEHFVIRIPDSLPLDGAGPLLCAGVTVYSPLRHFGL-DKPGMNIGVVGLG 649
Query: 194 GLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRSLDFILD 253
GLGHMAVKF KAFG VTVISTSP K+ EA E LGAD F++S +P+Q+QAA +L+ I+D
Sbjct: 650 GLGHMAVKFAKAFGAKVTVISTSPKKEKEAIEHLGADSFLVSRDPEQMQAATSTLNGIID 829
Query: 254 TVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIKETQEML 313
TVSA H ++P++ LLK NG L +VGA KPL+LP F L+ G++SI G +IGGIKETQEM+
Sbjct: 830 TVSASHPVVPLIGLLKSNGKLVMVGAVAKPLELPIFSLLGGRKSIAGSLIGGIKETQEMI 1009
Query: 314 DVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANA 356
D KHN+T +IE++ D +N A +RL+K DV+YRFVIDI N+
Sbjct: 1010DFAAKHNVTPEIEVVPIDYVNTAMERLVKGDVKYRFVIDIGNS 1138
>TC85885 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (97%)
Length = 1330
Score = 422 bits (1086), Expect = e-119
Identities = 201/348 (57%), Positives = 256/348 (72%)
Frame = +3
Query: 8 HTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPV 67
H + GWAA DSSG ++P+ F RRE G DV K+LYCGICHTD+H K++WG ++YP+
Sbjct: 93 HPKKAFGWAARDSSGVLSPFNFSRRETGEKDVAFKVLYCGICHTDLHMMKNEWGNSIYPL 272
Query: 68 VPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIF 127
VPGHE+ G++T+VGS VE FK GDKVGVG + SC CE C D ENYC + +
Sbjct: 273 VPGHELAGIVTEVGSKVEKFKVGDKVGVGYMVDSCRSCENCAEDLENYCPQQTVTCGAKY 452
Query: 128 WDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRI 187
DGS+TYGGYS +V D +V+ IP+SLP+D A PLLCAG+TV+SPL+ H + G I
Sbjct: 453 RDGSVTYGGYSDSMVADEHFVIRIPDSLPLDVAGPLLCAGVTVYSPLR-HFQLDKPGMNI 629
Query: 188 GVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRS 247
GVVGLGGLGHMAVKF KAFG +VTVISTSPSK+ EA E LGAD F++S +PDQ+QAA +
Sbjct: 630 GVVGLGGLGHMAVKFAKAFGANVTVISTSPSKEKEAIEHLGADSFLVSRDPDQMQAAMGT 809
Query: 248 LDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIK 307
L+ I+DTVSA H +LP++ LLK NG L +VG KPL+LP F L+ G++ + G +IGGIK
Sbjct: 810 LNGIIDTVSASHPILPLIGLLKSNGKLVMVGGVAKPLELPVFSLLGGRKLVAGSLIGGIK 989
Query: 308 ETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIAN 355
ETQEM+D +HN+T DIE++ D +N A +RL K DV+YRFVIDI N
Sbjct: 990 ETQEMIDFAAEHNVTPDIEVVPIDYVNTAMERLEKADVKYRFVIDIGN 1133
>BF645529 weakly similar to GP|22475166|gb putative sinapyl alcohol
dehydrogenase {Populus tremula x Populus tremuloides},
partial (40%)
Length = 658
Score = 372 bits (955), Expect(2) = e-113
Identities = 174/178 (97%), Positives = 176/178 (98%)
Frame = +3
Query: 23 KITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEITGVITKVGS 82
++TPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEITGVITKVGS
Sbjct: 96 QVTPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEITGVITKVGS 275
Query: 83 DVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSITYGGYSQMLV 142
DVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSITYGGYSQMLV
Sbjct: 276 DVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSITYGGYSQMLV 455
Query: 143 VDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLGGLGHMAV 200
VDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDH LVSTAGKRIGVVGLG LGHMAV
Sbjct: 456 VDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHXLVSTAGKRIGVVGLGXLGHMAV 629
Score = 54.7 bits (130), Expect(2) = e-113
Identities = 25/28 (89%), Positives = 27/28 (96%)
Frame = +1
Query: 1 MAKTTPNHTQTVSGWAAHDSSGKITPYT 28
MAKTTPNHTQTVSGWAAHDSSGK+ P+T
Sbjct: 31 MAKTTPNHTQTVSGWAAHDSSGKL-PHT 111
>TC77145 homologue to SP|P31656|CADH_MEDSA Cinnamyl-alcohol dehydrogenase
(EC 1.1.1.195) (CAD). [Alfalfa] {Medicago sativa},
complete
Length = 1384
Score = 350 bits (899), Expect = 4e-97
Identities = 171/344 (49%), Positives = 235/344 (67%)
Frame = +1
Query: 10 QTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVP 69
+T G AA D SG +TPYT+ R G DV IKI YCG+CH+D+H K+D G++ YP+VP
Sbjct: 76 RTTVGLAAKDPSGILTPYTYTLRNTGPDDVYIKIHYCGVCHSDLHQIKNDLGMSNYPMVP 255
Query: 70 GHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWD 129
GHE+ G + +VGS+V FK G+ VGVG L C C C ++ E YC K + YN ++ D
Sbjct: 256 GHEVVGEVLEVGSNVTRFKVGEIVGVGLLVGCCKSCRACDSEIEQYCNKKIWSYNDVYTD 435
Query: 130 GSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGV 189
G IT GG+++ VV+ ++VV IPE L + APLLCAG+TV+SPL GL T G R G+
Sbjct: 436 GKITQGGFAESTVVEQKFVVKIPEGLAPEQVAPLLCAGVTVYSPLSHFGL-KTPGLRGGI 612
Query: 190 VGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRSLD 249
+GLGG+GHM VK KAFGHHVTVIS+S K+ EA E LGAD +++S++ +Q A SLD
Sbjct: 613 LGLGGVGHMGVKVAKAFGHHVTVISSSDKKKKEALEDLGADSYLVSSDTVGMQEAADSLD 792
Query: 250 FILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIKET 309
+I+DTV H L P L LLK++G L ++G + PLQ ++ G++SI G +G +KET
Sbjct: 793 YIIDTVPVGHPLEPYLSLLKIDGKLILMGVINTPLQFVTPMVMLGRKSITGSFVGSVKET 972
Query: 310 QEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDI 353
+EML+ + ++ IE++ D IN+AF+RL KNDVRYRFV+D+
Sbjct: 973 EEMLEFWKEKGLSSMIEIVTMDYINKAFERLEKNDVRYRFVVDV 1104
>TC90155 similar to SP|Q9ZRF1|MTD_FRAAN Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (78%)
Length = 1051
Score = 309 bits (792), Expect = 9e-85
Identities = 154/280 (55%), Positives = 202/280 (72%)
Frame = +2
Query: 76 VITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSITYG 135
++T+VGS VE FK GDKVGVG L SC C+ C + ENYC K + DG++TYG
Sbjct: 8 IVTEVGSKVEKFKVGDKVGVGYLIDSCRSCQDCNDNLENYCPKFVVTCGAKYRDGTVTYG 187
Query: 136 GYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLGGL 195
GYS +V D +V+ IP+++P++ A PLLCAG+TV+SPL+ GL G IGVVGLGGL
Sbjct: 188 GYSDSMVADEHFVIRIPDNIPLEFAGPLLCAGVTVYSPLRFFGL-DKPGLHIGVVGLGGL 364
Query: 196 GHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRSLDFILDTV 255
GHMAVKF KAFG +VTVISTSP+K+ EA E LGAD F+IS++P Q+Q A +LD I+DTV
Sbjct: 365 GHMAVKFAKAFGANVTVISTSPNKEKEAIEHLGADSFLISSDPKQIQGAIGTLDGIIDTV 544
Query: 256 SADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIKETQEMLDV 315
SA H LLP++ LLK +G L ++G +PLQLP + LI G++ + G +GG+KETQEM++
Sbjct: 545 SAVHPLLPMIGLLKSHGKLIMLGVIVQPLQLPEYTLIQGRKILAGSQVGGLKETQEMINF 724
Query: 316 CGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIAN 355
+HN+ DIE++ D +N A QRL K DV+YRFVIDI N
Sbjct: 725 AAEHNVKPDIEVVPIDYVNTAMQRLAKGDVKYRFVIDIGN 844
>CA923178 weakly similar to SP|P93257|MTD_ Probable mannitol dehydrogenase
(EC 1.1.1.255) (NAD-dependent mannitol dehydrogenase).,
partial (34%)
Length = 668
Score = 276 bits (706), Expect = 9e-75
Identities = 140/140 (100%), Positives = 140/140 (100%)
Frame = -3
Query: 221 AEAKERLGADDFIISTNPDQLQAAKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAP 280
AEAKERLGADDFIISTNPDQLQAAKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAP
Sbjct: 666 AEAKERLGADDFIISTNPDQLQAAKRSLDFILDTVSADHALLPILELLKVNGTLFIVGAP 487
Query: 281 DKPLQLPAFPLIFGKRSIKGGIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRL 340
DKPLQLPAFPLIFGKRSIKGGIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRL
Sbjct: 486 DKPLQLPAFPLIFGKRSIKGGIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRL 307
Query: 341 LKNDVRYRFVIDIANASPAT 360
LKNDVRYRFVIDIANASPAT
Sbjct: 306 LKNDVRYRFVIDIANASPAT 247
>BG648660 similar to SP|Q9ZRF1|MTD_ Probable mannitol dehydrogenase (EC
1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Strawberry], partial (64%)
Length = 721
Score = 276 bits (706), Expect = 9e-75
Identities = 132/228 (57%), Positives = 166/228 (71%)
Frame = +1
Query: 8 HTQTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPV 67
H++ GWAA DSSG ++P+ F RRE DV +++LYCGICH+D+H AK++WG T YP+
Sbjct: 40 HSKKAFGWAARDSSGVLSPFNFSRREICEKDVALRVLYCGICHSDLHKAKNEWGTTNYPL 219
Query: 68 VPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIF 127
VPGHEI G++T+VGS V+ FK GD+VGVGCL SC C+ C + ENYC +L +
Sbjct: 220 VPGHEIVGIVTEVGSKVKKFKVGDRVGVGCLVDSCHSCQNCVDNLENYCPQLTLTDGSKY 399
Query: 128 WDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRI 187
DG+ T+GG+S +V D +V IP+ LP+DAAAPLLCAGITV+SPL+ GL I
Sbjct: 400 SDGTSTHGGFSDSMVSDEHFVFRIPDQLPLDAAAPLLCAGITVYSPLRHFGL-DKPDMNI 576
Query: 188 GVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIIS 235
GVVGLGGLGHMAVKF KAFG +VTVISTSP K+ EA E L AD F+IS
Sbjct: 577 GVVGLGGLGHMAVKFAKAFGANVTVISTSPKKENEALEHLRADSFLIS 720
>TC80850 homologue to SP|O82515|MTD_MEDSA Probable mannitol dehydrogenase
(EC 1.1.1.255) (NAD-dependent mannitol dehydrogenase).
[Alfalfa], partial (61%)
Length = 710
Score = 272 bits (696), Expect = 1e-73
Identities = 126/207 (60%), Positives = 159/207 (75%)
Frame = +1
Query: 14 GWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEI 73
GWAA D+SG ++P+ F RRENG DV++KILYCG+CH+D+H K+DWG T YPVVPGHEI
Sbjct: 94 GWAARDTSGTLSPFHFSRRENGDDDVSVKILYCGVCHSDLHTLKNDWGFTTYPVVPGHEI 273
Query: 74 TGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSIT 133
GV+TKVG +V+ F+ GD VGVG + SC CE C D E YC KL F YN + G+ T
Sbjct: 274 VGVVTKVGINVKKFRVGDNVGVGVIVESCQTCENCNQDLEQYCPKLVFTYNSPY-KGTRT 450
Query: 134 YGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLG 193
+GGYS +VV RYVV P++LP+DA APLLCAGITV+SP+K +G+ + GK +GV GLG
Sbjct: 451 HGGYSDFVVVHQRYVVQFPDNLPLDAGAPLLCAGITVYSPMKYYGM-TEPGKHLGVAGLG 627
Query: 194 GLGHMAVKFGKAFGHHVTVISTSPSKQ 220
GLGH+A+KFGKAFG VTVI+TSP+K+
Sbjct: 628 GLGHVAIKFGKAFGLKVTVITTSPNKE 708
>TC90520 weakly similar to PIR|T08581|T08581 cinnamyl-alcohol dehydrogenase
(EC 1.1.1.195) CAD1 - Arabidopsis thaliana, partial
(41%)
Length = 618
Score = 246 bits (627), Expect(2) = 2e-67
Identities = 127/189 (67%), Positives = 138/189 (72%), Gaps = 5/189 (2%)
Frame = +2
Query: 10 QTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVP 69
+ VSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVP
Sbjct: 65 KXVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVP 244
Query: 70 GHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIF-- 127
GHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQ F
Sbjct: 245 GHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQCCIQWYFLG 424
Query: 128 WDGSITY--GGYSQMLVVDYRYV-VHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAG 184
W + + + L Y+++ H+ L + PLKDHG
Sbjct: 425 W*HHLWWLLTNVGRRLQKAYQWMQQHLYSVLE*QYS-----------XPLKDHGFGINGR 571
Query: 185 KRIGVVGLG 193
++IGVVG G
Sbjct: 572 EKIGVVGFG 598
Score = 74.3 bits (181), Expect = 7e-14
Identities = 43/78 (55%), Positives = 50/78 (63%)
Frame = +3
Query: 122 VYNGIFWDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVS 181
VYNGIFWDGSITYGGYSQMLVVDYR + S LC ++ S + LVS
Sbjct: 402 VYNGIFWDGSITYGGYSQMLVVDYRKPTNGCSS------TSTLCWNNSIXSL*RTTVLVS 563
Query: 182 TAGKRIGVVGLGGLGHMA 199
TAGK++ + LGGLG MA
Sbjct: 564 TAGKKLVXLVLGGLGXMA 617
Score = 27.7 bits (60), Expect(2) = 2e-67
Identities = 11/11 (100%), Positives = 11/11 (100%)
Frame = +1
Query: 1 MAKTTPNHTQT 11
MAKTTPNHTQT
Sbjct: 37 MAKTTPNHTQT 69
>TC78752 similar to GP|22857590|gb|AAN09864.1 putative cinnamyl-alcohol
dehydrogenase {Oryza sativa (japonica cultivar-group)},
partial (50%)
Length = 668
Score = 209 bits (532), Expect = 1e-54
Identities = 104/188 (55%), Positives = 132/188 (69%), Gaps = 1/188 (0%)
Frame = +2
Query: 15 WAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEIT 74
WAA D+SG ++PY F RRE G DV +KI +CG+C+ DV AK+ G + YPVVPGHEI
Sbjct: 80 WAARDASGVLSPYKFNRRELGSEDVYVKITHCGVCYADVIWAKNKHGDSKYPVVPGHEIA 259
Query: 75 GVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEK-LQFVYNGIFWDGSIT 133
GV+ KVG +V+ FK GD VGVG SC +CEYC E +C K + +NG+ +DG+IT
Sbjct: 260 GVVAKVGPNVQRFKVGDHVGVGTYINSCRECEYCNDRFEVHCVKGSVYTFNGVDYDGTIT 439
Query: 134 YGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLG 193
GGYS +VV RY IP+S P+ +A PLLCAGITV+SP+ H + + GK +GVVGLG
Sbjct: 440 KGGYSTSIVVHERYCFLIPKSHPLASAGPLLCAGITVYSPMIRHNM-NQPGKSLGVVGLG 616
Query: 194 GLGHMAVK 201
GLGHMAVK
Sbjct: 617 GLGHMAVK 640
>TC78753 similar to PIR|E96751|E96751 hypothetical protein F28P22.13
[imported] - Arabidopsis thaliana, partial (47%)
Length = 865
Score = 177 bits (448), Expect = 7e-45
Identities = 92/169 (54%), Positives = 116/169 (68%)
Frame = +3
Query: 188 GVVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQAAKRS 247
GVVGLGGLGHMAVKFGKAFG VTV STS SK+ EA LGAD F++S+N + ++A +S
Sbjct: 3 GVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSLLGADQFVVSSNQEDMRALAKS 182
Query: 248 LDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFPLIFGKRSIKGGIIGGIK 307
LDFI+DT S DH P + LLK++G L +VG P + PA L G R++ G + GG K
Sbjct: 183 LDFIIDTASGDHLFDPYMSLLKISGVLVLVGFPSEVKFSPA-SLNLGSRTVAGSVTGGTK 359
Query: 308 ETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDIANA 356
E QEM+D C + I DIELI NEA +R++ DV+YRFVIDI N+
Sbjct: 360 EIQEMVDFCAANGIHPDIELIPIGYSNEALERVVNKDVKYRFVIDIENS 506
>BI263584 similar to SP|Q9P6C8|ADH1 Alcohol dehydrogenase I (EC 1.1.1.1).
{Neurospora crassa}, partial (57%)
Length = 682
Score = 122 bits (307), Expect = 2e-28
Identities = 68/177 (38%), Positives = 102/177 (57%), Gaps = 2/177 (1%)
Frame = +2
Query: 35 GGTDVTIKILYCGICHTDVHHAKDDWGI-TMYPVVPGHEITGVITKVGSDVEGFKDGDKV 93
G +V + + + G+CHTD+H + DW + T P+V GHE GV+ G V+ K G+KV
Sbjct: 158 GPDEVLVNVKFSGVCHTDLHAWQGDWPLDTKLPLVGGHEGAGVVVARGDLVKDVKIGEKV 337
Query: 94 GVGCLAASCLDCEYCKTDQENYCEKLQFVYNGIFWDGSITYGGYSQMLVVDYRYVVHIPE 153
G+ L SCL C YC+ E+ C + + +G DGS + Q + +V IPE
Sbjct: 338 GIKWLNGSCLSCSYCQNADESLC--AEALLSGYTVDGS-----FQQYAIAKAIHVARIPE 496
Query: 154 SLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGL-GGLGHMAVKFGKAFGHH 209
+++ +P+LCAGITV+ LK+ G+ AG+ I +VG GGLG +A ++ KA G H
Sbjct: 497 ECDLESISPILCAGITVYKGLKESGV--KAGQSIAIVGAGGGLGSIAXQYCKAMGIH 661
>TC79080 similar to GP|10177043|dbj|BAB10455. alcohol dehydrogenase-like
protein {Arabidopsis thaliana}, partial (96%)
Length = 1524
Score = 91.7 bits (226), Expect = 4e-19
Identities = 68/268 (25%), Positives = 113/268 (41%), Gaps = 27/268 (10%)
Frame = +3
Query: 38 DVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEITGVITKVGSD-----VEGFKDGDK 92
++ IK CG+CH+D+H K + + P V GHEITG + + G +E G +
Sbjct: 309 ELLIKTKGCGVCHSDLHVMKGEIPFSS-PCVVGHEITGEVVEHGQHTDSKTIERLPIGSR 485
Query: 93 VGVGCLAASCLDCEYCKTDQENYCEKLQFVYN---GIFWDGSI--------------TYG 135
V VG C +C YC ++ CE F YN G +DG + G
Sbjct: 486 V-VGAFIMPCGNCSYCSKGHDDLCEAF-FAYNRAKGTLYDGETRLFLRGSGNPIYMYSMG 659
Query: 136 GYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLGGL 195
G ++ VV + +P S+P +A L CA T + + H G + V+G GG+
Sbjct: 660 GLAEYCVVPANALAVLPNSMPYTESAILGCAVFTAYGAMA-HAAEVRPGDTVAVIGTGGV 836
Query: 196 GHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQLQ-----AAKRSLDF 250
G ++ +AFG + ++ E + LGA I S D ++ + +D
Sbjct: 837 GSSCLQIARAFGASDIIAVDVQDEKLEKAKTLGATHTINSAKEDPIEKILEITGGKGVDV 1016
Query: 251 ILDTVSADHALLPILELLKVNGTLFIVG 278
++ + + +K G ++G
Sbjct: 1017AVEALGRPQTFAQCTQSVKDGGKAVMIG 1100
>TC85844 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 (EC
1.1.1.1). [Garden pea] {Pisum sativum}, complete
Length = 1502
Score = 89.7 bits (221), Expect = 2e-18
Identities = 78/337 (23%), Positives = 141/337 (41%), Gaps = 30/337 (8%)
Frame = +2
Query: 38 DVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGVGC 97
+V +KIL+ +CHTDV+ + ++P + GHE G++ +G V K GD +
Sbjct: 197 EVRLKILFTSLCHTDVYFWEAKGQTPLFPRIFGHEAGGIVESIGEGVTHLKPGDH-ALPV 373
Query: 98 LAASCLDCEYCKTDQENYCEKL---------------QFVYNGIFWDGSITYGGYSQMLV 142
C DC +CK+++ N C+ L +F G + +S+ V
Sbjct: 374 FTGECGDCPHCKSEESNMCDLLRINTDRGVMINDNQSRFSLKGQPIHHFVGTSTFSEYTV 553
Query: 143 VDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLGGLGHMAVKF 202
V V I P+D L C GI + G + + GLG +G A +
Sbjct: 554 VHAGCVAKINPDAPLDKVCILSC-GICTGLGATINVAKPKPGSSVAIFGLGAVGLAAAEG 730
Query: 203 GKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQ------LQAAKRSLDFILDTVS 256
+ G + S + E ++ G ++F+ + D+ + +D ++
Sbjct: 731 ARISGASRIIGVDLVSSRFELAKKFGVNEFVNPKDHDKPVQQVIAEMTNGGVDRAVECTG 910
Query: 257 ADHALLPILELLKVN-GTLFIVGAPDKPLQLPAFPL-IFGKRSIKGGIIGGIKETQEMLD 314
+ A++ E + G +VG P+K P+ + +R++KG G K ++ +
Sbjct: 911 SIQAMISAFECVHDGWGVAVLVGVPNKDDAFKTHPMNLLNERTLKGTFYGNYKPRTDLPN 1090
Query: 315 VCGKHNITCDIELIKADT-------INEAFQRLLKND 344
V K+ + ++EL K T IN+AF +LK +
Sbjct: 1091VVEKY-MKGELELEKFITHTIPFSEINKAFDYMLKGE 1198
>TC76794 similar to GP|7416846|dbj|BAA94084.1 NAD-dependent sorbitol
dehydrogenase {Prunus persica}, partial (93%)
Length = 1691
Score = 84.0 bits (206), Expect = 8e-17
Identities = 76/300 (25%), Positives = 131/300 (43%), Gaps = 19/300 (6%)
Frame = +2
Query: 10 QTVSGWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKD----DWGITMY 65
Q ++ W ++ KI P+ G DV IK+ GIC +DVH+ K D+ I
Sbjct: 257 QNMAAWLVGLNTLKIQPFNLPSL--GPHDVRIKMKAVGICGSDVHYLKTLRCADF-IVKE 427
Query: 66 PVVPGHEITGVITKVGSDVEGFKDGDKVGVGCLAASCLDCEYCKTDQENYCEKLQFVYNG 125
P+V GHE G+I +VGS V+ GD+V + SC C++CK + N C ++
Sbjct: 428 PMVIGHECAGIIEEVGSQVKTLVPGDRVAIE-PGISCWRCDHCKLGRYNLCPDMK----- 589
Query: 126 IFWDGSITYGGYSQMLVVDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGK 185
F+ +G + +V +PE++ ++ A +C ++V G+ +
Sbjct: 590 -FFATPPVHGSLANQIVHPADLCFKLPENVSLEEGA--MCEPLSV-------GVHACRRA 739
Query: 186 RIG------VVGLGGLGHMAVKFGKAFGHHVTVISTSPSKQAEAKERLGADDFI-ISTN- 237
IG ++G G +G + + +AFG V+ + + LGADD + +STN
Sbjct: 740 NIGPETNVLIMGAGPIGLVTMLSARAFGAPRIVVVDVDDHRLSVAKSLGADDIVKVSTNI 919
Query: 238 PDQLQAAKR-------SLDFILDTVSADHALLPILELLKVNGTLFIVGAPDKPLQLPAFP 290
D + K+ +D D + + L + G + +VG + +P P
Sbjct: 920 QDVAEEVKQIHNVLGAGVDVTFDCAGFNKTMTTALTATQPGGKVCLVGMGHSEMTVPLTP 1099
>BQ158078 similar to GP|10187159|emb cDNA~Strawberry alcohol dehydrogenase
{Fragaria x ananassa}, partial (19%)
Length = 970
Score = 83.2 bits (204), Expect = 1e-16
Identities = 34/60 (56%), Positives = 44/60 (72%)
Frame = +2
Query: 14 GWAAHDSSGKITPYTFKRRENGGTDVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEI 73
GWAA D SG ++P+ F RRE G DVT K+LY GICH+D+H AK++WG YP+ GHE+
Sbjct: 26 GWAARDPSGVLSPFNFSRRETGEKDVTFKVLY*GICHSDLHMAKNEWGTYFYPLDSGHEL 205
>TC78975 similar to GP|21592656|gb|AAM64605.1 alcohol dehydrogenase
putative {Arabidopsis thaliana}, partial (77%)
Length = 1691
Score = 82.0 bits (201), Expect = 3e-16
Identities = 84/337 (24%), Positives = 140/337 (40%), Gaps = 33/337 (9%)
Frame = +2
Query: 38 DVTIKILYCGICHTDVHHAKDDWGITM-YPVVPGHEITGVITKVGSDVEGFKDGDKVGVG 96
+V IKILY ICHTD+ K + YP + GHE +G++ VG V K+ DKV V
Sbjct: 209 EVRIKILYTSICHTDLSGWKGECEPQRAYPRIFGHEASGIVESVGEGVNDMKENDKV-VL 385
Query: 97 CLAASCLDCEYCKTDQENYCEKLQF-VYNGIFWDGSITYGG--------------YSQML 141
C +C+ C + N CEK + DG+ + +++
Sbjct: 386 IFNGECGECKCCMCKKTNMCEKSGVDPMKKLMCDGTSRFSSIDGKPIFHFLNTSTFTEYT 565
Query: 142 VVDYRYVVHI--PESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLGGLGHMA 199
VVD V + ++L + L C T ++ V AG + + GLG +G
Sbjct: 566 VVDSACAVKLNTEDNLSLKKLTLLSCGVSTGIGAAWNNANVH-AGSSVAIFGLGAVGLAV 742
Query: 200 VKFGKAFG-HHVTVISTSPSKQAEAKERLGADDFIISTNPDQ------LQAAKRSLDFIL 252
+ +A G + + +P K E +G +FI + ++ + +D+
Sbjct: 743 AEGARARGASKIIGVDINPDKFNNTAETMGITEFINPKDEEKPVYEIIREMTDGGVDYSF 922
Query: 253 DTVSADHALL-PILELLKVNGTLFIVGAPDKPLQLPAFPL-IFGKRSIKGGIIGGIKETQ 310
+ + L L + + G ++G P LP P+ +F R I+G + GG K
Sbjct: 923 ECTGNLNVLRDSFLSVHEGWGLTVLLGIHGSPKLLPIHPMELFDGRRIEGSVFGGFKGKS 1102
Query: 311 EMLDV---CGKHNITCD---IELIKADTINEAFQRLL 341
++ ++ C K I D + D IN+AF L+
Sbjct: 1103QLPNLATECMKGAIKLDNFITHELPFDEINQAFNLLI 1213
>TC85846 homologue to SP|P12886|ADH1_PEA Alcohol dehydrogenase 1 (EC
1.1.1.1). [Garden pea] {Pisum sativum}, partial (78%)
Length = 955
Score = 72.8 bits (177), Expect = 2e-13
Identities = 60/265 (22%), Positives = 106/265 (39%), Gaps = 22/265 (8%)
Frame = +3
Query: 38 DVTIKILYCGICHTDVHHAKDDWGITMYPVVPGHEITGVITKVGSDVEGFKDGDKVGVGC 97
+V +KIL+ +CHTDV+ + ++P + GHE G++ VG V K GD +
Sbjct: 153 EVRLKILFTSLCHTDVYFWEAKGQTPLFPRIFGHEAGGIVESVGEGVTHLKPGDH-ALPV 329
Query: 98 LAASCLDCEYCKTDQENYCEKL---------------QFVYNGIFWDGSITYGGYSQMLV 142
C DC +CK+++ N C+ L +F G + +S+ V
Sbjct: 330 FTGECGDCPHCKSEESNMCDLLRINTDRGVMINDNQSRFSIKGQPIHHFVGTSTFSEYTV 509
Query: 143 VDYRYVVHIPESLPMDAAAPLLCAGITVFSPLKDHGLVSTAGKRIGVVGLGGLGHMAVKF 202
V V I P+D L C GI + G + + GLG +G A +
Sbjct: 510 VHAGCVAKINPDAPLDKVCILSC-GICTGLGATINVAKPKPGSSVAIFGLGAVGLAAAEG 686
Query: 203 GKAFGHHVTVISTSPSKQAEAKERLGADDFIISTNPDQ------LQAAKRSLDFILDTVS 256
+ G + S + E ++ G ++F+ + D+ + +D ++
Sbjct: 687 ARISGASRIIGVDLVSSRFELAKKFGVNEFVNPKDHDKPVQQVIAEMTNGGVDRAVECTG 866
Query: 257 ADHALLPILELLKVN-GTLFIVGAP 280
+ A++ E + G +VG P
Sbjct: 867 SIQAMISAFECVHDGWGVAVLVGVP 941
>BQ146920 similar to GP|8099340|gb|A ELI3 {Lycopersicon esculentum}, partial
(20%)
Length = 686
Score = 72.4 bits (176), Expect = 3e-13
Identities = 36/62 (58%), Positives = 44/62 (70%)
Frame = +2
Query: 294 GKRSIKGGIIGGIKETQEMLDVCGKHNITCDIELIKADTINEAFQRLLKNDVRYRFVIDI 353
G+R G IGG+KETQ+MLD KHN+ IE+I D +N A +RLLK DV+YRFVIDI
Sbjct: 8 GER**PGSNIGGMKETQDMLDFAAKHNVKPTIEVIPMDYVNTAMERLLKADVKYRFVIDI 187
Query: 354 AN 355
N
Sbjct: 188 GN 193
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,283,440
Number of Sequences: 36976
Number of extensions: 161256
Number of successful extensions: 766
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 729
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 733
length of query: 360
length of database: 9,014,727
effective HSP length: 97
effective length of query: 263
effective length of database: 5,428,055
effective search space: 1427578465
effective search space used: 1427578465
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146790.6


