

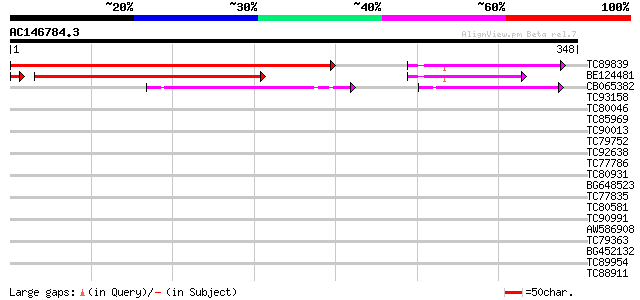
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146784.3 - phase: 0
(348 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC ... 398 e-111
BE124481 285 2e-77
CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fra... 75 3e-14
TC93158 similar to PIR|T01123|T01123 hypothetical protein At2g32... 36 0.025
TC80046 similar to SP|P22699|GUN6_DICDI Endoglucanase precursor ... 35 0.043
TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsi... 35 0.043
TC90013 homologue to PIR|JN0688|JN0688 histone H4 - sea squirt (... 33 0.16
TC79752 similar to GP|11994670|dbj|BAB02898. gene_id:K7P8.17~unk... 33 0.16
TC92638 similar to GP|23463525|gb|AAN33391.1 conserved hypotheti... 30 0.19
TC77786 33 0.21
TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem tra... 32 0.36
BG648523 similar to GP|9955540|emb| putative protein {Arabidopsi... 32 0.47
TC77835 similar to GP|20197423|gb|AAM15069.1 unknown protein {Ar... 32 0.47
TC80581 similar to GP|19683021|gb|AAL92644.1 hypothetical protei... 31 0.62
TC90991 homologue to PIR|A25642|HSZM4 histone H4 - maize, complete 31 0.62
AW586908 weakly similar to GP|2104814|emb| pleiotropic effects o... 31 0.81
TC79363 similar to GP|12322790|gb|AAG51387.1 unknown protein; 10... 31 0.81
BG452132 weakly similar to PIR|C85343|C853 hypothetical protein ... 31 0.81
TC89954 PIR|A25642|HSZM4 histone H4 - maize, complete 30 1.1
TC88911 weakly similar to GP|13926280|gb|AAK49610.1 AT5g14920/F2... 30 1.1
>TC89839 homologue to SP|P23466|CYAA_SACKL Adenylate cyclase (EC 4.6.1.1)
(ATP pyrophosphate-lyase) (Adenylyl cyclase). [Yeast],
partial (0%)
Length = 665
Score = 398 bits (1023), Expect = e-111
Identities = 193/200 (96%), Positives = 195/200 (97%)
Frame = +3
Query: 1 MSSSKFGSHMQTLVQTGNMDQLEQEVQELRGEVTTLRAEVEKLASLVSSLMATNDPPLVQ 60
MSSSKFGSHMQTLVQT MDQLEQEV ELRGEVTTLRAEVEKL SLVSSLMATNDPPLVQ
Sbjct: 63 MSSSKFGSHMQTLVQTQKMDQLEQEVHELRGEVTTLRAEVEKLTSLVSSLMATNDPPLVQ 242
Query: 61 QRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILL 120
QRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIP+KYADLLPILL
Sbjct: 243 QRPQSPYQPMCPQKPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPVKYADLLPILL 422
Query: 121 KKNLVQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFD 180
KKNL+QTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFD
Sbjct: 423 KKNLIQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFD 602
Query: 181 DQDIKVLLQQQHLAPHSVAA 200
DQDIKVLLQQQHLAPHS AA
Sbjct: 603 DQDIKVLLQQQHLAPHSGAA 662
Score = 106 bits (264), Expect = 2e-23
Identities = 56/118 (47%), Positives = 71/118 (59%), Gaps = 21/118 (17%)
Frame = +3
Query: 245 YQPQFQQYQQQPRQQAPRIKF---------------------DPIPMKYGELFPYLLERN 283
YQP Q +PRQQAPR DPIP+KY +L P LL++N
Sbjct: 261 YQPMCPQ---KPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPVKYADLLPILLKKN 431
Query: 284 LVQTRPPPPIPKKLPARWRPDLFCVFHQGAQGHDVERCFSLKIEVQKLIEDDLIPFEE 341
L+QT P P +P LP +RPDL CVFHQGA GHD E+C+ LK EVQKLIE+++ F++
Sbjct: 432 LIQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDD 605
>BE124481
Length = 538
Score = 285 bits (729), Expect(2) = 2e-77
Identities = 136/142 (95%), Positives = 138/142 (96%)
Frame = +3
Query: 16 TGNMDQLEQEVQELRGEVTTLRAEVEKLASLVSSLMATNDPPLVQQRPQSPYQPMCPQKP 75
T MDQLEQEV ELRGEVTTLRAEVEKL SLVSSLMATNDPPLVQQRPQSPYQPMCPQKP
Sbjct: 111 TQKMDQLEQEVHELRGEVTTLRAEVEKLTSLVSSLMATNDPPLVQQRPQSPYQPMCPQKP 290
Query: 76 RQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLVQTLPLPRVPN 135
RQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIP+KYADLLPILLKKNL+QTLPLPRVPN
Sbjct: 291 RQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPVKYADLLPILLKKNLIQTLPLPRVPN 470
Query: 136 SLPPWYRPDLNCVFHQGAPGHD 157
SLPPWYRPDLNCVFHQGAPGHD
Sbjct: 471 SLPPWYRPDLNCVFHQGAPGHD 536
Score = 81.6 bits (200), Expect = 4e-16
Identities = 44/94 (46%), Positives = 52/94 (54%), Gaps = 21/94 (22%)
Frame = +3
Query: 245 YQPQFQQYQQQPRQQAPRIKF---------------------DPIPMKYGELFPYLLERN 283
YQP Q +PRQQAPR DPIP+KY +L P LL++N
Sbjct: 264 YQPMCPQ---KPRQQAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPVKYADLLPILLKKN 434
Query: 284 LVQTRPPPPIPKKLPARWRPDLFCVFHQGAQGHD 317
L+QT P P +P LP +RPDL CVFHQGA GHD
Sbjct: 435 LIQTLPLPRVPNSLPPWYRPDLNCVFHQGAPGHD 536
Score = 22.3 bits (46), Expect(2) = 2e-77
Identities = 9/9 (100%), Positives = 9/9 (100%)
Frame = +2
Query: 1 MSSSKFGSH 9
MSSSKFGSH
Sbjct: 83 MSSSKFGSH 109
>CB065382 similar to PIR|T09671|T096 RPE15 protein - alfalfa (fragment),
partial (9%)
Length = 624
Score = 75.5 bits (184), Expect = 3e-14
Identities = 45/89 (50%), Positives = 52/89 (57%)
Frame = -2
Query: 252 YQQQPRQQAPRIKFDPIPMKYGELFPYLLERNLVQTRPPPPIPKKLPARWRPDLFCVFHQ 311
+QQ+ QQA R F IPM Y E P LL R R P P LP R+R DL C FHQ
Sbjct: 512 HQQRQ*QQA-RPTFPLIPMLYAE*LPTLLLRGHCTIRQGKPPPDPLPPRFRSDLKCDFHQ 336
Query: 312 GAQGHDVERCFSLKIEVQKLIEDDLIPFE 340
GA GHDVE C++LK V+KLI + FE
Sbjct: 335 GALGHDVEGCYALKHIVKKLINQGKLTFE 249
Score = 70.1 bits (170), Expect = 1e-12
Identities = 53/129 (41%), Positives = 64/129 (49%), Gaps = 1/129 (0%)
Frame = -2
Query: 85 PQNQVPQKFIPQNQVQKASQCDP-IPMKYADLLPILLKKNLVQTLPLPRVPNSLPPWYRP 143
P+N P Q Q Q+A P IPM YA+ LP LL + P+ LPP +R
Sbjct: 536 PRNNHPPVH-QQRQ*QQARPTFPLIPMLYAE*LPTLLLRGHCTIRQGKPPPDPLPPRFRS 360
Query: 144 DLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDDQDIKVLLQQQHLAPHSVAAVRP 203
DL C FHQGA GHD E CY LK V+KLI +F++ VL L H AAV
Sbjct: 359 DLKCDFHQGALGHDVEGCYALKHIVKKLINQGKLTFENNVPHVL--DNPLPNH--AAVNM 192
Query: 204 ITNVVQDPG 212
I + PG
Sbjct: 191 IEVYEEAPG 165
>TC93158 similar to PIR|T01123|T01123 hypothetical protein At2g32840
[imported] - Arabidopsis thaliana, partial (9%)
Length = 673
Score = 35.8 bits (81), Expect = 0.025
Identities = 27/86 (31%), Positives = 39/86 (44%), Gaps = 1/86 (1%)
Frame = +2
Query: 235 PIMNVVRNPGYQP-QFQQYQQQPRQQAPRIKFDPIPMKYGELFPYLLERNLVQTRPPPPI 293
P N NP +P + Q QQQP+QQA ++ P P Y P N PPP
Sbjct: 86 PFTNPNPNPNSKPIHYPQQQQQPQQQALHVR-SPNPFLYPFASPSRASANHAVGGYPPPP 262
Query: 294 PKKLPARWRPDLFCVFHQGAQGHDVE 319
P P++ +P L G +G +++
Sbjct: 263 P---PSQPQPPLLYSHGGGVRGMNLD 331
>TC80046 similar to SP|P22699|GUN6_DICDI Endoglucanase precursor (EC
3.2.1.4) (Endo-1 4-beta-glucanase) (Spore germination
protein 270-6), partial (11%)
Length = 643
Score = 35.0 bits (79), Expect = 0.043
Identities = 39/116 (33%), Positives = 51/116 (43%), Gaps = 12/116 (10%)
Frame = +1
Query: 51 MATNDPPLVQQRPQSPYQ-------PMCPQKPRQ---QAPRQSIPQNQVPQKFIPQNQVQ 100
+ T PL Q+ PQ P Q PQKP Q Q P Q +PQ + PQK +PQ Q
Sbjct: 250 LQTRSLPLPQRPPQRPPQRPPQKPPQRPPQKPPQRPPQRPPQKLPQ-RPPQK-LPQRPPQ 423
Query: 101 KASQCDPIPMKYADLLPILLKKNLVQTLPLPRVP--NSLPPWYRPDLNCVFHQGAP 154
K Q +P K +K ++ LP P+ P LP +P L + H P
Sbjct: 424 KLPQ--KLPQKPPQRPLPHPQKPPLRPLPHPQKPPLRPLPHPQKPPLRPLPHPQKP 585
>TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsis thaliana},
partial (31%)
Length = 2756
Score = 35.0 bits (79), Expect = 0.043
Identities = 26/67 (38%), Positives = 31/67 (45%), Gaps = 2/67 (2%)
Frame = +2
Query: 195 PHSVAAVRPITNVVQDPGY--QPQFRPSQQQYPDSHFVTTILPIMNVVRNPGYQPQFQQY 252
PHS V+ VQDPGY Q QF QQQ + + P +QPQ Q
Sbjct: 1568 PHSRIQVQ---QHVQDPGYLLQQQFELQQQQQQ--------FELQQQQQQPQHQPQQSQP 1714
Query: 253 QQQPRQQ 259
Q QP+QQ
Sbjct: 1715 QHQPQQQ 1735
>TC90013 homologue to PIR|JN0688|JN0688 histone H4 - sea squirt (Styela
plicata), complete
Length = 574
Score = 33.1 bits (74), Expect = 0.16
Identities = 19/54 (35%), Positives = 28/54 (51%), Gaps = 5/54 (9%)
Frame = -3
Query: 261 PRIKFDPIPMKY-----GELFPYLLERNLVQTRPPPPIPKKLPARWRPDLFCVF 309
P I+F P P++ G + P +L R +++ R PP P P RPD+F F
Sbjct: 200 PEIRFTP-PLRARRLIAGLVIP*MLSRRILR*RFAPPFPSPFPPLPRPDIFLCF 42
>TC79752 similar to GP|11994670|dbj|BAB02898. gene_id:K7P8.17~unknown
protein {Arabidopsis thaliana}, partial (2%)
Length = 2137
Score = 33.1 bits (74), Expect = 0.16
Identities = 50/240 (20%), Positives = 80/240 (32%), Gaps = 3/240 (1%)
Frame = +1
Query: 21 QLEQEVQELRGEVTTLRAE-VEKLASLVSSLMATNDPPLVQQRPQSPYQP--MCPQKPRQ 77
Q + V EL +VT ++ + + + S+ + PP VQQ P Q + P
Sbjct: 517 QRQMMVPELPMQVTQGNSQGIPAFSGMSSAFNSQTTPPSVQQYPVHAQQQSHLSNPHPHL 696
Query: 78 QAPRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLPILLKKNLVQTLPLPRVPNSL 137
Q P Q Q + + Q+Q+ Q + + L+ Q P P
Sbjct: 697 QGPNHPNNQQQAYIRLAKERQLQQQQQQRYLQQQQLSATNALIPHVQAQAQPPISSPQQN 876
Query: 138 PPWYRPDLNCVFHQGAPGHDTEQCYPLKEEVQKLIENNVWSFDDQDIKVLLQQQHLAPHS 197
+P + +P + P+ + Q QQ+H P
Sbjct: 877 SSQAQPQNSSQQVSVSPATPSSPLTPMSSQHQ-------------------QQKHDLPQP 999
Query: 198 VAAVRPITNVVQDPGYQPQFRPSQQQYPDSHFVTTILPIMNVVRNPGYQPQFQQYQQQPR 257
+ P ++V Q Q + Q+QY S R QPQ Q QQQ +
Sbjct: 1000GFSRNPGSSVTSQAVKQRQRQAQQRQYQQS------------ARQHPNQPQHAQAQQQAK 1143
>TC92638 similar to GP|23463525|gb|AAN33391.1 conserved hypothetical protein
{Brucella suis 1330}, partial (6%)
Length = 612
Score = 29.6 bits (65), Expect(2) = 0.19
Identities = 11/13 (84%), Positives = 13/13 (99%)
Frame = +1
Query: 106 DPIPMKYADLLPI 118
DPIPMKYA+LLP+
Sbjct: 262 DPIPMKYAELLPV 300
Score = 21.9 bits (45), Expect(2) = 0.19
Identities = 9/19 (47%), Positives = 13/19 (68%)
Frame = +3
Query: 86 QNQVPQKFIPQNQVQKASQ 104
+ Q PQ+FI Q+ VQK +
Sbjct: 207 RQQAPQQFISQD*VQKTQR 263
>TC77786
Length = 1094
Score = 32.7 bits (73), Expect = 0.21
Identities = 12/27 (44%), Positives = 20/27 (73%)
Frame = +3
Query: 147 CVFHQGAPGHDTEQCYPLKEEVQKLIE 173
C +H+ GHDTE+C+ L+E ++ LI+
Sbjct: 825 CCYHRSR-GHDTEECFQLREAIEDLIK 902
Score = 29.6 bits (65), Expect = 1.8
Identities = 11/27 (40%), Positives = 19/27 (69%)
Frame = +3
Query: 307 CVFHQGAQGHDVERCFSLKIEVQKLIE 333
C +H+ ++GHD E CF L+ ++ LI+
Sbjct: 825 CCYHR-SRGHDTEECFQLREAIEDLIK 902
>TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem transcription
factor M1 {Apium graveolens}, partial (67%)
Length = 1193
Score = 32.0 bits (71), Expect = 0.36
Identities = 21/52 (40%), Positives = 26/52 (49%), Gaps = 3/52 (5%)
Frame = +2
Query: 56 PPLVQQRPQSPYQPMCPQKPRQQAP---RQSIPQNQVPQKFIPQNQVQKASQ 104
P L QQ Q Q PQ + Q +Q IPQ Q Q+ +PQ Q Q+ SQ
Sbjct: 560 PQLQQQMQQQQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQQLPQLQQQQLSQ 715
>BG648523 similar to GP|9955540|emb| putative protein {Arabidopsis thaliana},
partial (9%)
Length = 770
Score = 31.6 bits (70), Expect = 0.47
Identities = 28/84 (33%), Positives = 35/84 (41%), Gaps = 5/84 (5%)
Frame = +3
Query: 60 QQRPQSPYQPMCPQKPRQQA--PRQSIPQNQVPQKFIPQNQVQKASQCDPIPMKYADLLP 117
Q P S P P P P Q P Q PQ+ IPQ+Q Q P+PM +P
Sbjct: 381 QPHPGSIGAPPLPPGPHPSLLNPNQQQPFQQNPQQ-IPQHQQQLQQHMGPLPM--PPNMP 551
Query: 118 ILLKKNLVQTLP---LPRVPNSLP 138
+ + LP LPR P +P
Sbjct: 552 QIQHSSHSSMLPHQHLPRPPPQMP 623
>TC77835 similar to GP|20197423|gb|AAM15069.1 unknown protein {Arabidopsis
thaliana}, partial (87%)
Length = 691
Score = 31.6 bits (70), Expect = 0.47
Identities = 20/51 (39%), Positives = 22/51 (42%), Gaps = 2/51 (3%)
Frame = +3
Query: 211 PGYQPQFRPSQQQYPDSHFVTTILPIMNVVRNPGYQPQF--QQYQQQPRQQ 259
PGY PQ P QQ YP + GY PQ+ Q Q PRQQ
Sbjct: 180 PGYPPQGYPPQQGYPPQGYPP----------QQGYPPQYAPQYAQPPPRQQ 302
>TC80581 similar to GP|19683021|gb|AAL92644.1 hypothetical protein
{Dictyostelium discoideum}, partial (5%)
Length = 850
Score = 31.2 bits (69), Expect = 0.62
Identities = 24/88 (27%), Positives = 44/88 (49%), Gaps = 5/88 (5%)
Frame = +1
Query: 21 QLEQEVQELRGEVTTLRAEVEKLASLVSSLMATNDP--PLVQQRPQSPYQPMCPQKPRQQ 78
QL Q+ +L+ + L+ ++ A L + + N PL+ Q+P+S QP+ Q+ +Q
Sbjct: 331 QLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQL 510
Query: 79 APRQSIPQNQ---VPQKFIPQNQVQKAS 103
+Q Q Q + Q+ Q+Q+ S
Sbjct: 511 LQQQLQLQQQYQRLQQQLASSAQLQQNS 594
>TC90991 homologue to PIR|A25642|HSZM4 histone H4 - maize, complete
Length = 517
Score = 31.2 bits (69), Expect = 0.62
Identities = 13/33 (39%), Positives = 20/33 (60%)
Frame = -2
Query: 273 GELFPYLLERNLVQTRPPPPIPKKLPARWRPDL 305
G + P++L R+ ++ R PP PK P RPD+
Sbjct: 141 GFVIPWMLSRSTLRCRFAPPFPKPFPPFPRPDI 43
>AW586908 weakly similar to GP|2104814|emb| pleiotropic effects on cellular
differentiation and slug behaviour {Dictyostelium
discoideum}, partial (8%)
Length = 668
Score = 30.8 bits (68), Expect = 0.81
Identities = 32/106 (30%), Positives = 49/106 (46%), Gaps = 2/106 (1%)
Frame = +2
Query: 21 QLEQEVQELRG-EVTTLRAEVEKLASLVSSLMATNDPPLVQQRPQSPYQPMCPQKPRQQA 79
Q +Q++Q L + L+ + ++ L S N P QQ+PQ Q PQ+P+QQ
Sbjct: 35 QQQQKMQSLLPMPLDQLQQQQQRQQQLAGS---QNLPQPQQQQPQLTQQS--PQQPQQQQ 199
Query: 80 PRQSIPQNQVPQK-FIPQNQVQKASQCDPIPMKYADLLPILLKKNL 124
Q QN I NQ+ +QC P+ Y+ L LL ++
Sbjct: 200 QHQQPCQNTTMNNGTIGSNQI--PNQCVQQPVTYSQLQQQLLSGSM 331
>TC79363 similar to GP|12322790|gb|AAG51387.1 unknown protein; 108050-105786
{Arabidopsis thaliana}, partial (32%)
Length = 976
Score = 30.8 bits (68), Expect = 0.81
Identities = 12/32 (37%), Positives = 20/32 (62%)
Frame = +1
Query: 46 LVSSLMATNDPPLVQQRPQSPYQPMCPQKPRQ 77
L+ L+ ++DPP+V Q P++ +C KP Q
Sbjct: 349 LLQLLLKSDDPPIVTQSEMDPHRRICNPKPPQ 444
>BG452132 weakly similar to PIR|C85343|C853 hypothetical protein AT4g29420
[imported] - Arabidopsis thaliana, partial (19%)
Length = 577
Score = 30.8 bits (68), Expect = 0.81
Identities = 17/48 (35%), Positives = 20/48 (41%)
Frame = +1
Query: 215 PQFRPSQQQYPDSHFVTTILPIMNVVRNPGYQPQFQQYQQQPRQQAPR 262
PQ R + + DSH LP RN FQ QQPR P+
Sbjct: 106 PQLRFLRSSFVDSHLFHVSLPQFTFPRNQAPHHPFQNRFQQPRSPFPQ 249
>TC89954 PIR|A25642|HSZM4 histone H4 - maize, complete
Length = 509
Score = 30.4 bits (67), Expect = 1.1
Identities = 13/34 (38%), Positives = 20/34 (58%)
Frame = -2
Query: 273 GELFPYLLERNLVQTRPPPPIPKKLPARWRPDLF 306
G + P++L R+ ++ PP P LP RPD+F
Sbjct: 136 GFVIPWMLSRSTLRCLFAPPFPNPLPPFPRPDIF 35
Score = 30.4 bits (67), Expect = 1.1
Identities = 12/31 (38%), Positives = 21/31 (67%)
Frame = -2
Query: 115 LLPILLKKNLVQTLPLPRVPNSLPPWYRPDL 145
++P +L ++ ++ L P PN LPP+ RPD+
Sbjct: 130 VIPWMLSRSTLRCLFAPPFPNPLPPFPRPDI 38
>TC88911 weakly similar to GP|13926280|gb|AAK49610.1 AT5g14920/F2G14_40
{Arabidopsis thaliana}, partial (11%)
Length = 1320
Score = 30.4 bits (67), Expect = 1.1
Identities = 27/112 (24%), Positives = 47/112 (41%), Gaps = 10/112 (8%)
Frame = +2
Query: 38 AEVEKLASLVSSLMATNDPPLVQQRPQSPYQPMCPQKPRQQAPRQS-----IPQNQVPQK 92
A + L V++ A P+V + P+ P P+ AP+ S +P+++ P
Sbjct: 167 APAKPLVPKVTTPTAAPVKPIVPKTTTPTSAPLKPPVPKAPAPKPSPVKPPVPKSKPPTT 346
Query: 93 FIPQNQVQKASQCDPIPMKYADLLPILLKKN---LVQTLPLP--RVPNSLPP 139
++ V + P+ + + P L K +Q P P + P SLPP
Sbjct: 347 APVKSPVPDPPKLAPVKLPVPKVTPALSPKTPSPKIQPPPHPPKKAPVSLPP 502
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,254,293
Number of Sequences: 36976
Number of extensions: 207642
Number of successful extensions: 1410
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 1288
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1379
length of query: 348
length of database: 9,014,727
effective HSP length: 97
effective length of query: 251
effective length of database: 5,428,055
effective search space: 1362441805
effective search space used: 1362441805
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146784.3


