

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146784.1 - phase: 0
(151 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
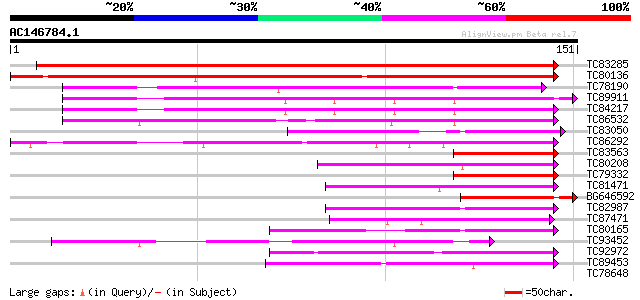
TC83285 weakly similar to GP|7406995|gb|AAF61863.1| DNA-binding ... 282 4e-77
TC80136 similar to PIR|T50861|T50861 DNA-binding protein 4 [impo... 250 1e-67
TC78190 weakly similar to PIR|T49041|T49041 DNA-binding protein-... 79 9e-16
TC89911 similar to GP|13507101|gb|AAK28442.1 WRKY DNA-binding pr... 59 7e-10
TC84217 similar to GP|13507101|gb|AAK28442.1 WRKY DNA-binding pr... 55 1e-08
TC86532 similar to GP|17064164|gb|AAL35289.1 WRKY transcription ... 51 2e-07
TC83050 similar to SP|Q9FGZ4|WR48_ARATH Probable WRKY transcript... 49 6e-07
TC86292 similar to GP|17064164|gb|AAL35289.1 WRKY transcription ... 49 1e-06
TC83563 similar to PIR|T52092|T52092 DNA-binding protein WRKY2 [... 43 4e-05
TC80208 similar to GP|17064158|gb|AAL35286.1 WRKY transcription ... 42 7e-05
TC79332 similar to PIR|JC6203|JC6203 SP8 binding protein homolog... 42 9e-05
TC81471 similar to PIR|A84913|A84913 probable WRKY-type DNA bind... 42 1e-04
BG646592 similar to GP|13507101|gb| WRKY DNA-binding protein 53 ... 41 2e-04
TC82987 similar to GP|15991740|gb|AAL13047.1 WRKY transcription ... 41 2e-04
TC87471 SP|Q9LP56|WR65_ARATH Probable WRKY transcription factor ... 41 2e-04
TC80165 similar to PIR|A84913|A84913 probable WRKY-type DNA bind... 40 3e-04
TC93452 similar to PIR|D84902|D84902 probable WRKY-type DNA bind... 40 4e-04
TC92972 similar to GP|9187622|emb|CAB97004.1 WRKY DNA binding pr... 39 6e-04
TC89453 similar to GP|9187622|emb|CAB97004.1 WRKY DNA binding pr... 39 6e-04
TC78648 similar to PIR|S51529|S51529 SPF1 protein - sweet potato... 39 0.001
>TC83285 weakly similar to GP|7406995|gb|AAF61863.1| DNA-binding protein 3
{Nicotiana tabacum}, partial (17%)
Length = 657
Score = 282 bits (721), Expect = 4e-77
Identities = 137/139 (98%), Positives = 137/139 (98%)
Frame = +3
Query: 8 HGSGRGKVIDELRRGRELANQLKNILNESGDFDDINGSTTPFAEDLLKEVLTTFTNSLLF 67
HGSGRGKVIDELRRGRELANQLKNILNESGDFDDINGSTTPFAEDLLKEVLTTFTNSLLF
Sbjct: 81 HGSGRGKVIDELRRGRELANQLKNILNESGDFDDINGSTTPFAEDLLKEVLTTFTNSLLF 260
Query: 68 LNNNPTSEGSDMQLTKSEDSLESNCKSTSIVKERRGCYKRRKVSQTWEKESDRPVEDGHQ 127
LNNNPTSEGSDMQLTKSEDSLESNCKSTSIVKERRGCYKRRKVSQTWEKESDRPVEDGHQ
Sbjct: 261 LNNNPTSEGSDMQLTKSEDSLESNCKSTSIVKERRGCYKRRKVSQTWEKESDRPVEDGHQ 440
Query: 128 WRKYGQKKILHTDFPRYIY 146
WRKYGQKKILHTDFPR Y
Sbjct: 441 WRKYGQKKILHTDFPRNYY 497
>TC80136 similar to PIR|T50861|T50861 DNA-binding protein 4 [imported] -
common tobacco (fragment), partial (33%)
Length = 1276
Score = 250 bits (639), Expect = 1e-67
Identities = 127/147 (86%), Positives = 136/147 (92%), Gaps = 1/147 (0%)
Frame = +2
Query: 1 MKNENLAHGSGRGKVIDELRRGRELANQLKNILNESGDFDDINGSTTP-FAEDLLKEVLT 59
M+NENLAHG GRGKV+DEL RGRELAN+L+NILNESGD + +GST P FAEDLLKEVLT
Sbjct: 68 MENENLAHG-GRGKVVDELLRGRELANKLRNILNESGDIYNNDGSTIPPFAEDLLKEVLT 244
Query: 60 TFTNSLLFLNNNPTSEGSDMQLTKSEDSLESNCKSTSIVKERRGCYKRRKVSQTWEKESD 119
TFTNSLLFLNNNPTSEGSDMQLTKSEDSLESNCKS SIVKERRGCYKRRK+SQTWEKES+
Sbjct: 245 TFTNSLLFLNNNPTSEGSDMQLTKSEDSLESNCKS-SIVKERRGCYKRRKISQTWEKESE 421
Query: 120 RPVEDGHQWRKYGQKKILHTDFPRYIY 146
+P EDGHQWRKYGQKKILHTDFPR Y
Sbjct: 422 QPEEDGHQWRKYGQKKILHTDFPRNYY 502
>TC78190 weakly similar to PIR|T49041|T49041 DNA-binding protein-like -
Arabidopsis thaliana, partial (27%)
Length = 1254
Score = 78.6 bits (192), Expect = 9e-16
Identities = 48/137 (35%), Positives = 77/137 (56%), Gaps = 8/137 (5%)
Frame = +1
Query: 15 VIDELRRGRELANQLKNILNESGDFDDINGSTTPFAEDLLKEVLTTFTNSLLFLNN---- 70
+I EL +G+E A QLK +L + + +P ++L+ VL +F+ ++ +N+
Sbjct: 199 LIKELLQGKEYATQLKLLLK-----NPVCSDGSPSVKELVTNVLRSFSETISVMNSTEDC 363
Query: 71 ----NPTSEGSDMQLTKSEDSLESNCKSTSIVKERRGCYKRRKVSQTWEKESDRPVEDGH 126
N + + KSED+ ES + + K+RRG YKRRK +T S + + D H
Sbjct: 364 SLDVNDSGSLVEADDLKSEDTSESKKRLSPTTKDRRGSYKRRKTDETRTIVS-KTIGDTH 540
Query: 127 QWRKYGQKKILHTDFPR 143
WRKYGQK+IL+++FPR
Sbjct: 541 SWRKYGQKEILNSNFPR 591
>TC89911 similar to GP|13507101|gb|AAK28442.1 WRKY DNA-binding protein 53
{Arabidopsis thaliana}, partial (21%)
Length = 1302
Score = 58.9 bits (141), Expect = 7e-10
Identities = 50/162 (30%), Positives = 71/162 (42%), Gaps = 25/162 (15%)
Frame = +1
Query: 15 VIDELRRGRELANQLKNILNESGDFDDINGSTTPFAEDLLKEVLTTFTNSLLFLNNNP-- 72
+IDEL G ELA QL+ LN ++ S+ E L++++++TF +L +N
Sbjct: 25 LIDELLHGLELARQLQIYLN-------VSSSSNETREILIQKIISTFEKALEMVNRKRHV 183
Query: 73 -TSEGSDMQLTKSE--------DSLESNCKSTSIVKER------RGCYKRRKVSQTWEKE 117
E S Q + DS +C S +R KR + W K
Sbjct: 184 NMGESSKQQYPSASGTLAIRISDSPPLSCSPRSEDSDRDFNRDHNASRKRNTLPIRWTKH 363
Query: 118 --------SDRPVEDGHQWRKYGQKKILHTDFPRYIYFYEFN 151
+ P++DG+ WRKYGQK IL PRYI EFN
Sbjct: 364 IRVEPGMAMEGPLDDGYSWRKYGQKDILGAMHPRYI-ISEFN 486
>TC84217 similar to GP|13507101|gb|AAK28442.1 WRKY DNA-binding protein 53
{Arabidopsis thaliana}, partial (21%)
Length = 706
Score = 54.7 bits (130), Expect = 1e-08
Identities = 46/157 (29%), Positives = 67/157 (42%), Gaps = 25/157 (15%)
Frame = +1
Query: 15 VIDELRRGRELANQLKNILNESGDFDDINGSTTPFAEDLLKEVLTTFTNSLLFLNNNP-- 72
+IDEL G ELA QL+ LN ++ S+ E L++++++TF +L +N
Sbjct: 55 LIDELLHGLELARQLQIYLN-------VSSSSNETREILIQKIISTFEKALEMVNRKRHV 213
Query: 73 -TSEGSDMQLTKSE--------DSLESNCKSTSIVKER------RGCYKRRKVSQTWEKE 117
E S Q + DS +C S +R KR + W K
Sbjct: 214 NMGESSKQQYPSASGTLAIRISDSPPLSCSPRSEDSDRDFNRDHNASRKRNTLPIRWTKH 393
Query: 118 --------SDRPVEDGHQWRKYGQKKILHTDFPRYIY 146
+ P++DG+ WRKYGQK IL PR Y
Sbjct: 394 IRVEPGMAMEGPLDDGYSWRKYGQKDILGAMHPRGYY 504
>TC86532 similar to GP|17064164|gb|AAL35289.1 WRKY transcription factor 41
{Arabidopsis thaliana}, partial (25%)
Length = 1620
Score = 51.2 bits (121), Expect = 2e-07
Identities = 41/153 (26%), Positives = 70/153 (44%), Gaps = 21/153 (13%)
Frame = +1
Query: 15 VIDELRRGRELANQLKNIL------NESGDFDDINGSTTPFAEDLLKEVLTTFTNSLLFL 68
+I EL +G ELA +L + + + S + ++T + L+ +++ + ++
Sbjct: 265 LIGELIQGEELAKKLYDQIFSPSSPSSSTSSSSSSSTSTSSHQVLIDKLILSIETAITMA 444
Query: 69 NNNPTSEGSDMQLTKSEDSLESNCKSTSIVKE-------RRGCYKRRKVSQTWEKE---- 117
N EG T + + ++S+C + S E + K+RK W ++
Sbjct: 445 KGN---EGKIK--TNNVNMMDSHCSNGSPKSEVQDSKFKHKHVSKKRKTVPKWTEQVKVY 609
Query: 118 ----SDRPVEDGHQWRKYGQKKILHTDFPRYIY 146
++ VEDGH WRKYGQK IL FPR Y
Sbjct: 610 SGTAAEGSVEDGHSWRKYGQKDILGAKFPRGYY 708
>TC83050 similar to SP|Q9FGZ4|WR48_ARATH Probable WRKY transcription factor
48 (WRKY DNA-binding protein 48). [Mouse-ear cress],
partial (31%)
Length = 974
Score = 49.3 bits (116), Expect = 6e-07
Identities = 26/74 (35%), Positives = 40/74 (53%)
Frame = +2
Query: 75 EGSDMQLTKSEDSLESNCKSTSIVKERRGCYKRRKVSQTWEKESDRPVEDGHQWRKYGQK 134
EG D K++ L+ K+ +E R + + E D ++DG++WRKYGQK
Sbjct: 443 EGDDQDQDKTKKQLKPKKKNQKKEREPRFAFMTKS-------EVDH-LDDGYRWRKYGQK 598
Query: 135 KILHTDFPRYIYFY 148
+ ++ FPRYIY Y
Sbjct: 599 AVKNSPFPRYIYIY 640
>TC86292 similar to GP|17064164|gb|AAL35289.1 WRKY transcription factor 41
{Arabidopsis thaliana}, partial (21%)
Length = 1529
Score = 48.5 bits (114), Expect = 1e-06
Identities = 48/172 (27%), Positives = 80/172 (45%), Gaps = 26/172 (15%)
Frame = +3
Query: 1 MKNE-NLAHGSGRGKVIDELRRGRELANQLKNILNESGDFDDINGSTTPFA----EDLLK 55
M+NE N H + +I+EL +G ++A +LK L TP++ + ++
Sbjct: 222 MENECNWEHNT----LINELIQGMDVAKRLKEDLR------------TPYSLGTRDSQMQ 353
Query: 56 EVLTTFTNSLLFLNNNPTSEGSDMQLTKSEDSLESNCKSTS---IVKERRGCY------- 105
+L+++ +L L N ++ S +T SL + ST+ + ++ G +
Sbjct: 354 MILSSYEKALQILRCNESTSKSQ-PVTPVITSLPESPVSTNGSPLSEDFDGTFQDHQEVK 530
Query: 106 ---KRRKVSQTW--------EKESDRPVEDGHQWRKYGQKKILHTDFPRYIY 146
K+RKV+ W + + P EDG+ WRKYGQK IL T PR Y
Sbjct: 531 NKSKKRKVAPKWMDQIRVSCDSSLEGPNEDGYNWRKYGQKDILGTKHPRSYY 686
>TC83563 similar to PIR|T52092|T52092 DNA-binding protein WRKY2 [imported] -
common tobacco, partial (21%)
Length = 838
Score = 43.1 bits (100), Expect = 4e-05
Identities = 15/28 (53%), Positives = 22/28 (78%)
Frame = +1
Query: 119 DRPVEDGHQWRKYGQKKILHTDFPRYIY 146
D+P +DG+ WRKYGQK++ ++FPR Y
Sbjct: 634 DKPADDGYNWRKYGQKQVKGSEFPRSYY 717
>TC80208 similar to GP|17064158|gb|AAL35286.1 WRKY transcription factor 28
{Arabidopsis thaliana}, partial (31%)
Length = 1294
Score = 42.4 bits (98), Expect = 7e-05
Identities = 21/65 (32%), Positives = 39/65 (59%), Gaps = 1/65 (1%)
Frame = +1
Query: 83 KSEDSLESNCKSTSIVKERRGCYKRRKVSQTWEKESD-RPVEDGHQWRKYGQKKILHTDF 141
K E ES+ + V +++G K+++ + +S+ +EDG++WRKYGQK + ++ +
Sbjct: 484 KEEPGQESSKNGNNKVDKKKGEKKQKEPRFAFMTKSEVDQLEDGYRWRKYGQKAVKNSPY 663
Query: 142 PRYIY 146
PR Y
Sbjct: 664 PRSYY 678
>TC79332 similar to PIR|JC6203|JC6203 SP8 binding protein homolog -
cucumber, partial (49%)
Length = 1486
Score = 42.0 bits (97), Expect = 9e-05
Identities = 14/28 (50%), Positives = 22/28 (78%)
Frame = +2
Query: 119 DRPVEDGHQWRKYGQKKILHTDFPRYIY 146
D+P +DG+ WRKYGQK++ +++PR Y
Sbjct: 713 DKPADDGYNWRKYGQKQVKGSEYPRSYY 796
Score = 29.3 bits (64), Expect = 0.63
Identities = 10/21 (47%), Positives = 15/21 (70%)
Frame = +3
Query: 122 VEDGHQWRKYGQKKILHTDFP 142
++DG++WRKYGQK + P
Sbjct: 1269 LDDGYRWRKYGQKVVKGNPHP 1331
>TC81471 similar to PIR|A84913|A84913 probable WRKY-type DNA binding protein
[imported] - Arabidopsis thaliana, partial (43%)
Length = 1274
Score = 41.6 bits (96), Expect = 1e-04
Identities = 22/64 (34%), Positives = 35/64 (54%), Gaps = 2/64 (3%)
Frame = +2
Query: 85 EDSLESNCKSTSIVKERRGCYKRRKVSQT--WEKESDRPVEDGHQWRKYGQKKILHTDFP 142
ED E K+ +K ++ KR++ + K +EDG++WRKYGQK + ++ FP
Sbjct: 437 EDEEEEKQKTNKQLKTKKTNLKRQREPRFAFMTKSEVDHLEDGYRWRKYGQKAVKNSPFP 616
Query: 143 RYIY 146
R Y
Sbjct: 617 RSYY 628
>BG646592 similar to GP|13507101|gb| WRKY DNA-binding protein 53 {Arabidopsis
thaliana}, partial (19%)
Length = 638
Score = 41.2 bits (95), Expect = 2e-04
Identities = 19/31 (61%), Positives = 22/31 (70%)
Frame = +2
Query: 121 PVEDGHQWRKYGQKKILHTDFPRYIYFYEFN 151
P++DG+ WRKYGQK IL PRYI EFN
Sbjct: 278 PLDDGYSWRKYGQKDILGAMHPRYI-ISEFN 367
>TC82987 similar to GP|15991740|gb|AAL13047.1 WRKY transcription factor 71
{Arabidopsis thaliana}, partial (33%)
Length = 975
Score = 40.8 bits (94), Expect = 2e-04
Identities = 20/62 (32%), Positives = 34/62 (54%)
Frame = +1
Query: 85 EDSLESNCKSTSIVKERRGCYKRRKVSQTWEKESDRPVEDGHQWRKYGQKKILHTDFPRY 144
ED + K + K++ K + + + E D +EDG++WRKYGQK + ++ +PR
Sbjct: 568 EDGDDEKSKKENKAKKKEKKVKEPRFAFLTKSEIDH-LEDGYRWRKYGQKAVKNSPYPRS 744
Query: 145 IY 146
Y
Sbjct: 745 YY 750
>TC87471 SP|Q9LP56|WR65_ARATH Probable WRKY transcription factor 65 (WRKY
DNA-binding protein 65). [Mouse-ear cress], partial (9%)
Length = 578
Score = 40.8 bits (94), Expect = 2e-04
Identities = 24/63 (38%), Positives = 36/63 (57%), Gaps = 3/63 (4%)
Frame = +1
Query: 86 DSLESNCKSTSIVK--ERRGCYKRR-KVSQTWEKESDRPVEDGHQWRKYGQKKILHTDFP 142
DS+ SN + +S+ +RRG KR ++ + ++ P D WRKYGQK I + +P
Sbjct: 262 DSIPSNMERSSLSSSAKRRGIKKRVVQIPMKNVEGTNTPPSDSWAWRKYGQKPIKGSPYP 441
Query: 143 RYI 145
RYI
Sbjct: 442 RYI 450
>TC80165 similar to PIR|A84913|A84913 probable WRKY-type DNA binding protein
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1013
Score = 40.4 bits (93), Expect = 3e-04
Identities = 23/77 (29%), Positives = 41/77 (52%)
Frame = +1
Query: 70 NNPTSEGSDMQLTKSEDSLESNCKSTSIVKERRGCYKRRKVSQTWEKESDRPVEDGHQWR 129
+N T + ++ QL K + ++N K + +++ + E D +EDG++WR
Sbjct: 508 HNKTVDQTNNQLNKQLKAKKTNQKKP----------REARIAFMTKSEVDH-LEDGYRWR 654
Query: 130 KYGQKKILHTDFPRYIY 146
KYGQK + ++ FPR Y
Sbjct: 655 KYGQKAVKNSPFPRSYY 705
>TC93452 similar to PIR|D84902|D84902 probable WRKY-type DNA binding protein
[imported] - Arabidopsis thaliana, partial (10%)
Length = 488
Score = 40.0 bits (92), Expect = 4e-04
Identities = 32/126 (25%), Positives = 59/126 (46%), Gaps = 8/126 (6%)
Frame = +1
Query: 12 RGKVIDELRRGRELANQLKNIL----NESGDFDDINGSTTPFAEDLLKEVLTTFTNSLLF 67
R +I+EL G+ELANQL N + NE+ +F L+ ++++T+ +L
Sbjct: 121 RDSIINELVHGKELANQLSNHIILSSNETNEF-------------LIDKIISTYQKALTM 261
Query: 68 LNNNPTSEGSDMQLTKSEDSLESNCKSTSIVKER----RGCYKRRKVSQTWEKESDRPVE 123
LN D ++ S S + + ++ ++ +K+RK W+++S E
Sbjct: 262 LN------VGDGKMKDSHSSFTNESPKSEVIIDQEFNHNALFKKRKSMPKWKEQS----E 411
Query: 124 DGHQWR 129
D H+ R
Sbjct: 412 DLHKHR 429
Score = 26.2 bits (56), Expect = 5.3
Identities = 9/17 (52%), Positives = 13/17 (75%)
Frame = +2
Query: 118 SDRPVEDGHQWRKYGQK 134
S+ ++D + WRKYGQK
Sbjct: 434 SEGSLDDEYSWRKYGQK 484
>TC92972 similar to GP|9187622|emb|CAB97004.1 WRKY DNA binding protein
{Solanum tuberosum}, partial (46%)
Length = 672
Score = 39.3 bits (90), Expect = 6e-04
Identities = 25/77 (32%), Positives = 39/77 (50%)
Frame = +2
Query: 70 NNPTSEGSDMQLTKSEDSLESNCKSTSIVKERRGCYKRRKVSQTWEKESDRPVEDGHQWR 129
N GS++ + K + L++ K TS K + K + V QT + ++DG +WR
Sbjct: 80 NVKPGNGSNIHV-KIAEYLKTKKKETSSQKGGKEIMKHKYVFQT--RSQIDILDDGFRWR 250
Query: 130 KYGQKKILHTDFPRYIY 146
KYG+K + FPR Y
Sbjct: 251 KYGEKMVKDNKFPRSYY 301
>TC89453 similar to GP|9187622|emb|CAB97004.1 WRKY DNA binding protein
{Solanum tuberosum}, partial (56%)
Length = 701
Score = 39.3 bits (90), Expect = 6e-04
Identities = 24/79 (30%), Positives = 39/79 (48%), Gaps = 1/79 (1%)
Frame = +1
Query: 69 NNNPTSEGSDMQLTKSEDSLESNCKSTSIVKERRGCYKRRKVSQTWEKESDRPV-EDGHQ 127
NN + D + K E+ + VK ++G K +K ++ S + +DG++
Sbjct: 301 NNEISVTHDDHEDVKGEEGSVNVIIDQQDVK-KKGEKKAKKPKYAFQTRSQVDILDDGYR 477
Query: 128 WRKYGQKKILHTDFPRYIY 146
WRKYGQK + + FPR Y
Sbjct: 478 WRKYGQKAVKNNKFPRSYY 534
>TC78648 similar to PIR|S51529|S51529 SPF1 protein - sweet potato, partial
(18%)
Length = 990
Score = 38.5 bits (88), Expect = 0.001
Identities = 25/78 (32%), Positives = 39/78 (49%), Gaps = 4/78 (5%)
Frame = +1
Query: 73 TSEGSDMQLTKSEDS----LESNCKSTSIVKERRGCYKRRKVSQTWEKESDRPVEDGHQW 128
T+ S+ Q T+S S ++ S S+ G Y + + +E R EDG+ W
Sbjct: 616 TASKSEFQSTQSYSSEIVPIKPEIHSNSVTGS--GYYNYNNNASQFVREQKRS-EDGYNW 786
Query: 129 RKYGQKKILHTDFPRYIY 146
RKYGQK++ ++ PR Y
Sbjct: 787 RKYGQKQVKGSENPRSYY 840
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,550,225
Number of Sequences: 36976
Number of extensions: 55071
Number of successful extensions: 322
Number of sequences better than 10.0: 86
Number of HSP's better than 10.0 without gapping: 311
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 315
length of query: 151
length of database: 9,014,727
effective HSP length: 88
effective length of query: 63
effective length of database: 5,760,839
effective search space: 362932857
effective search space used: 362932857
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC146784.1


