

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146777.12 + phase: 0
(238 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
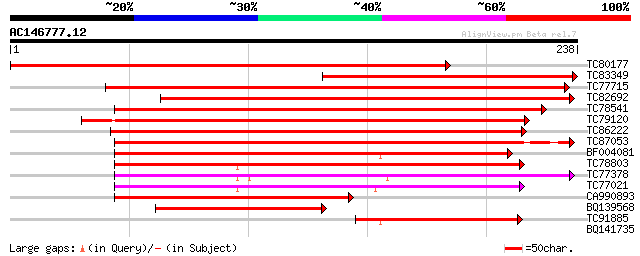
Score E
Sequences producing significant alignments: (bits) Value
TC80177 homologue to GP|9759372|dbj|BAB09831.1 ATP-dependent pro... 369 e-103
TC83349 similar to GP|9759372|dbj|BAB09831.1 ATP-dependent prote... 212 9e-56
TC77715 homologue to GP|15485610|emb|CAC67407. Clp protease 2 pr... 189 1e-48
TC82692 homologue to PIR|T52455|T52455 ATP-dependent clp protein... 173 6e-44
TC78541 similar to GP|13899067|gb|AAK48955.1 ATP-dependent Clp p... 169 7e-43
TC79120 similar to GP|14596089|gb|AAK68772.1 ATP-dependent Clp p... 156 6e-39
TC86222 similar to GP|9968665|emb|CAC06126.1 putative ATP-depend... 141 2e-34
TC87053 similar to PIR|T52454|T52454 ATP-dependent Clp proteinas... 137 5e-33
BF004081 similar to SP|P12210|CLPP ATP-dependent Clp protease pr... 135 2e-32
TC78803 similar to GP|19698857|gb|AAL91164.1 unknown protein {Ar... 135 2e-32
TC77378 similar to PIR|D86223|D86223 hypothetical protein [impor... 127 5e-30
TC77021 similar to GP|5360595|dbj|BAA82069.1 nClpP5 {Arabidopsis... 92 2e-19
CA990893 similar to SP|P30063|CLPP ATP-dependent Clp protease pr... 86 1e-17
BQ139568 similar to SP|P12210|CLPP_ ATP-dependent Clp protease p... 69 2e-12
TC91885 similar to SP|Q9BBQ9|CLPP_LOTJA ATP-dependent Clp protea... 58 3e-09
BQ141735 similar to PIR|S22697|S226 extensin - Volvox carteri (f... 27 9.1
>TC80177 homologue to GP|9759372|dbj|BAB09831.1 ATP-dependent protease
proteolytic subunit ClpP-like protein {Arabidopsis
thaliana}, partial (66%)
Length = 648
Score = 369 bits (947), Expect = e-103
Identities = 184/185 (99%), Positives = 184/185 (99%)
Frame = +2
Query: 1 MRGLFGITKTLFNGTKSSFSPCRNRAYSLIPMVIETSSRGERAYDIFSRLLKERIVCING 60
MRGLFGITKTLFNGTKSSFSPCRNRAYSLIPMVIETSSRGERAYDIFSRLLKERIVCING
Sbjct: 83 MRGLFGITKTLFNGTKSSFSPCRNRAYSLIPMVIETSSRGERAYDIFSRLLKERIVCING 262
Query: 61 PISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIYDTMQYIRSPINTICLG 120
PISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIYDTMQYIRSPINTICLG
Sbjct: 263 PISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIYDTMQYIRSPINTICLG 442
Query: 121 QAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAIHTKQIVRMWDALNELY 180
QAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAIHTKQIV MWDALNELY
Sbjct: 443 QAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAIHTKQIVXMWDALNELY 622
Query: 181 VKHTG 185
VKHTG
Sbjct: 623 VKHTG 637
>TC83349 similar to GP|9759372|dbj|BAB09831.1 ATP-dependent protease
proteolytic subunit ClpP-like protein {Arabidopsis
thaliana}, partial (43%)
Length = 814
Score = 212 bits (540), Expect = 9e-56
Identities = 105/107 (98%), Positives = 105/107 (98%)
Frame = +1
Query: 132 AGAKGQRRALPNATIMIHQPSGGYSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVI 191
AGAK Q RALPNATIMIHQPSGGYSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVI
Sbjct: 70 AGAKHQTRALPNATIMIHQPSGGYSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVI 249
Query: 192 QKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLVSDAVADEGKDKGSN 238
QKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLVSDAVADEGKDKGSN
Sbjct: 250 QKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLVSDAVADEGKDKGSN 390
>TC77715 homologue to GP|15485610|emb|CAC67407. Clp protease 2 proteolytic
subunit {Lycopersicon esculentum}, partial (76%)
Length = 1149
Score = 189 bits (479), Expect = 1e-48
Identities = 95/195 (48%), Positives = 136/195 (69%)
Frame = +3
Query: 41 ERAYDIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAG 100
ER + S+L + RI+ G + DD A+++V+QLL+L++ +P+K I MY+NSPGG+VTAG
Sbjct: 390 ERFQSVISQLFQYRIIRCGGAVDDDMANIIVSQLLYLDAVDPNKDIVMYVNSPGGSVTAG 569
Query: 101 LAIYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAK 160
+AI+DTM++IR ++T+C+G AASMG+ LL AG KG+R +LPN+ IMIHQP GG+ G
Sbjct: 570 MAIFDTMKHIRPDVSTVCVGLAASMGAFLLSAGTKGKRFSLPNSRIMIHQPLGGFQGGQT 749
Query: 161 DIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRP 220
DI I +++ LN TGQ L I ++ DRD+FM+A+EAKE+G+ID VI
Sbjct: 750 DIDIQANEMLHHKANLNGYLSYQTGQSLKKINQDTDRDFFMSAKEAKEYGLIDGVIMNPL 929
Query: 221 MTLVSDAVADEGKDK 235
L A A+EGKD+
Sbjct: 930 KALQPLATAEEGKDR 974
>TC82692 homologue to PIR|T52455|T52455 ATP-dependent clp proteinase (EC
3.4.21.-) chain P1 [imported] - Arabidopsis thaliana,
partial (73%)
Length = 1115
Score = 173 bits (438), Expect = 6e-44
Identities = 88/174 (50%), Positives = 123/174 (70%)
Frame = +2
Query: 64 DDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIYDTMQYIRSPINTICLGQAA 123
DD A+++V+QLL+L++ +PSK I MY+NSPGG+VTAG+AI+DTM++IR ++T+C+G AA
Sbjct: 449 DDMANIIVSQLLYLDAVDPSKDIVMYVNSPGGSVTAGMAIFDTMRHIRPDVSTVCVGLAA 628
Query: 124 SMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAIHTKQIVRMWDALNELYVKH 183
SMG+ LL AG KG+R +LPN+ IMIHQP GG+ G DI I +++ LN
Sbjct: 629 SMGAFLLSAGTKGKRYSLPNSRIMIHQPLGGFQGGQTDIDIQANEMLHHKANLNGYLSYQ 808
Query: 184 TGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLVSDAVADEGKDKGS 237
TGQ L+ I ++ DRD+FM+A+EAKE+G+ID VI L A A E D+ S
Sbjct: 809 TGQSLEKINQDTDRDFFMSAKEAKEYGLIDGVITNPLKALQPLASAAEVNDRAS 970
>TC78541 similar to GP|13899067|gb|AAK48955.1 ATP-dependent Clp protease;
nClpP3 {Arabidopsis thaliana}, partial (64%)
Length = 1158
Score = 169 bits (429), Expect = 7e-43
Identities = 79/181 (43%), Positives = 128/181 (70%)
Frame = +3
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
D + LL++RIV + + D TA V++QLLFL++++P+K I +++NSPGG+VTAG+ +Y
Sbjct: 285 DTTNMLLRQRIVFLGSQVDDMTADFVISQLLFLDADDPTKDIKLFINSPGGSVTAGMGVY 464
Query: 105 DTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAI 164
D M+ ++ ++T+CLG AASMG+ LL AG KG+R +PN+ +MIHQP G G+A +++I
Sbjct: 465 DAMKLCKADVSTVCLGLAASMGAFLLAAGTKGKRYCMPNSRVMIHQPLGSAGGKATEMSI 644
Query: 165 HTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLV 224
++++ +N++ + TG+P + I+ + DRD FM EAKE+G+ID+VID LV
Sbjct: 645 RARELMYHKIKINKILSRITGKPEEQIELDTDRDNFMNPWEAKEYGLIDDVIDDGKPGLV 824
Query: 225 S 225
+
Sbjct: 825 A 827
>TC79120 similar to GP|14596089|gb|AAK68772.1 ATP-dependent Clp
protease-like protein {Arabidopsis thaliana}, partial
(67%)
Length = 1110
Score = 156 bits (395), Expect = 6e-39
Identities = 79/188 (42%), Positives = 126/188 (67%)
Frame = +1
Query: 31 PMVIETSSRGERAYDIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYL 90
P E + RG + D LL+ERIV + I D A +++QLL L++++P+K I +++
Sbjct: 229 PQTPERAIRGAES-DTMGLLLRERIVFLGSEIDDFVADAIMSQLLLLDAQDPTKDIKLFI 405
Query: 91 NSPGGAVTAGLAIYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQ 150
NSPGG+++A +AIYD +Q +R+ ++TI +G +AS S++L G KG+R A+PN IMI Q
Sbjct: 406 NSPGGSLSATMAIYDAVQLVRADVSTIAMGISASTASIILGGGTKGKRLAMPNTRIMIAQ 585
Query: 151 PSGGYSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFG 210
P GG SGQA D+ I K+I+ + ++ + TG+ L+ + K++DRD +++ EA E+G
Sbjct: 586 PPGGASGQAIDVEIQAKEIMHNKNNVSRIISSFTGRSLEQVLKDIDRDRYLSPIEAVEYG 765
Query: 211 IIDEVIDQ 218
IID VID+
Sbjct: 766 IIDGVIDR 789
>TC86222 similar to GP|9968665|emb|CAC06126.1 putative ATP-dependent clp
serine protease proteolytic subunit {Pisum sativum},
complete
Length = 1054
Score = 141 bits (356), Expect = 2e-34
Identities = 70/175 (40%), Positives = 109/175 (62%)
Frame = +3
Query: 43 AYDIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLA 102
A D+ S L + RI+ I P++ A V++QL+ L + +P I MY+N PGG+ + LA
Sbjct: 357 ALDLSSVLFRNRIIFIGQPVNSQVAQRVISQLVTLATIDPDADILMYINCPGGSTYSVLA 536
Query: 103 IYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDI 162
IYD M +I+ + T+C G AAS G+LLL G KG R ++PNA IMI+QP G+ G +D+
Sbjct: 537 IYDCMSWIKPKVGTVCFGVAASQGTLLLAGGEKGMRYSMPNARIMINQPQCGFGGHVEDV 716
Query: 163 AIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVID 217
+ V+ + +++ TGQ L+ +Q+ +RD F++ EA EFG+ID V++
Sbjct: 717 RRLVNESVQSRYKMEKMFSAFTGQALEKVQEYTERDSFLSVSEALEFGLIDGVLE 881
>TC87053 similar to PIR|T52454|T52454 ATP-dependent Clp proteinase (EC
3.4.21.-) catalytic chain P2 [imported] - Arabidopsis
thaliana, partial (74%)
Length = 1135
Score = 137 bits (344), Expect = 5e-33
Identities = 72/193 (37%), Positives = 122/193 (62%)
Frame = +3
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
D+++ L +ER++ I I ++ ++ ++A LL+L+S + SK + +Y+N PGG +T +++Y
Sbjct: 372 DLWNALYRERVIFIGQEIDEELSNQILATLLYLDSVDNSKKLYLYINGPGGDLTPCMSLY 551
Query: 105 DTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYSGQAKDIAI 164
DTMQ +++PI T C+GQA + + LL AG KG R A+P A I+I P+G G+A DI
Sbjct: 552 DTMQSLQTPICTHCIGQAYGLAAFLLAAGEKGNRTAMPLARIVIQSPAGAARGRADDIQN 731
Query: 165 HTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVIDQRPMTLV 224
+++R+ D L TGQP++ I +++ R A+EA ++G+ID+++ RP +
Sbjct: 732 EAAELLRIRDYLFTELANKTGQPVEKITEDLKRVKRFDAQEALDYGLIDKIV--RPRRIK 905
Query: 225 SDAVADEGKDKGS 237
+DA KD GS
Sbjct: 906 ADA---PRKDAGS 935
>BF004081 similar to SP|P12210|CLPP ATP-dependent Clp protease proteolytic
subunit (EC 3.4.21.92) (Endopeptidase Clp). [Common
tobacco], partial (96%)
Length = 574
Score = 135 bits (339), Expect = 2e-32
Identities = 61/168 (36%), Positives = 114/168 (67%), Gaps = 1/168 (0%)
Frame = -2
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
D+++RL +ER++ + ++ + ++ +V +++L E+ ++ + +++NSPGG V +G+AI+
Sbjct: 504 DLYNRLFQERLLFLGQEVNTEISNQLVGLMVYLSLEDKTRDLYLFINSPGGEVISGMAIF 325
Query: 105 DTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGG-YSGQAKDIA 163
D MQ + + ++T+C+G AASMGSLLL G +R A P+A +MIHQP+ Y GQ +
Sbjct: 324 DIMQVVEAEVHTVCVGLAASMGSLLLVGGEFTKRLAFPHARVMIHQPATSLYEGQVGECM 145
Query: 164 IHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGI 211
+ +++++M + +Y + TG+ I ++M+RD FM+AEEA+ +GI
Sbjct: 144 LEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAEEAEAYGI 1
>TC78803 similar to GP|19698857|gb|AAL91164.1 unknown protein {Arabidopsis
thaliana}, partial (79%)
Length = 1200
Score = 135 bits (339), Expect = 2e-32
Identities = 74/180 (41%), Positives = 109/180 (60%), Gaps = 8/180 (4%)
Frame = +2
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPG--------GA 96
D+ S L K RIV + + ++VA+ L+L+ ++ +KPI MY+NS G G
Sbjct: 335 DLASYLYKNRIVYLGMSLVPSVTELMVAEFLYLQYDDETKPIYMYINSTGTTKGGEKLGY 514
Query: 97 VTAGLAIYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGYS 156
T LA+YD M+YI+ PI T+C+G A +LLL AGAKG R ALP++TIMI QP +
Sbjct: 515 ETEALAVYDLMRYIKPPIFTLCVGNAWGEAALLLAAGAKGNRSALPSSTIMIRQPIARFQ 694
Query: 157 GQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDEVI 216
GQA D+ + K+I R+ L L KHT + + I+ ++ R + + EA E+GIID+V+
Sbjct: 695 GQATDVNLARKEITRVKTELVNLLAKHTEKTPEQIEADIRRPKYFSPTEAVEYGIIDKVL 874
>TC77378 similar to PIR|D86223|D86223 hypothetical protein [imported] -
Arabidopsis thaliana, partial (54%)
Length = 1372
Score = 127 bits (318), Expect = 5e-30
Identities = 76/204 (37%), Positives = 113/204 (55%), Gaps = 11/204 (5%)
Frame = +1
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPG-----GAVTA 99
D+ S LL RIV I P+ +V+A+L+FL+ P +PI +Y+NS G G A
Sbjct: 469 DLPSLLLNGRIVYIGMPLVPAVTELVIAELMFLQWMAPKEPIYIYINSTGTTRADGETVA 648
Query: 100 ----GLAIYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPSGGY 155
G AIYD M +++ INT+ LG A LLL AG KG+R P+A MI QP
Sbjct: 649 MESEGFAIYDAMMQMKTEINTVALGAAVGQACLLLSAGTKGRRYMTPHAKAMIQQPRVPS 828
Query: 156 SG--QAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIID 213
SG A D+ IH K+++ D L +L KHT + + M R Y+M A AKEFG+ID
Sbjct: 829 SGLRPASDVLIHAKEVMVNRDTLVKLLAKHTENSEETVSNVMKRSYYMDALLAKEFGVID 1008
Query: 214 EVIDQRPMTLVSDAVADEGKDKGS 237
+++ + +++D + + ++ G+
Sbjct: 1009KILWRGQEKIMADVPSRDDRENGT 1080
>TC77021 similar to GP|5360595|dbj|BAA82069.1 nClpP5 {Arabidopsis thaliana},
partial (65%)
Length = 1521
Score = 91.7 bits (226), Expect = 2e-19
Identities = 54/182 (29%), Positives = 97/182 (52%), Gaps = 10/182 (5%)
Frame = +1
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPG---------G 95
D+ S LL RI + PI +++AQL++L+ +NP+KP+ +Y+NS G G
Sbjct: 487 DLPSLLLDSRICYLGMPIVPAVTELILAQLMWLDYDNPAKPVYVYINSSGTQNEKNETVG 666
Query: 96 AVTAGLAIYDTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNATIMIHQPS-GG 154
+ T +I D + +I+S + T+ L A +++L G KG R LP+++ + P
Sbjct: 667 SETDAYSIADMISHIKSDVYTVNLAMAYGQAAMILSLGKKGYRAVLPHSSAKVFLPKVHR 846
Query: 155 YSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAEEAKEFGIIDE 214
SG D+ I K++ + EL K TG+ + I K++ R + ++A E+G+ D+
Sbjct: 847 SSGSVADMWIKAKELEANSEYYIELLAKGTGKSKEEIAKDVQRTRYFQPQDAIEYGLADK 1026
Query: 215 VI 216
++
Sbjct: 1027IM 1032
>CA990893 similar to SP|P30063|CLPP ATP-dependent Clp protease proteolytic
subunit (EC 3.4.21.92) (Endopeptidase Clp).
[Beechdrops], partial (61%)
Length = 464
Score = 86.3 bits (212), Expect = 1e-17
Identities = 37/100 (37%), Positives = 71/100 (71%)
Frame = +1
Query: 45 DIFSRLLKERIVCINGPISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIY 104
D+++RL +ER++ + ++ + ++ +V +++L E+ ++ + +++NSPGG V +G+AI+
Sbjct: 133 DLYNRLFQERLLFLGQEVNTEISNQLVGLMVYLSLEDNTRDLYLFINSPGGEVISGMAIF 312
Query: 105 DTMQYIRSPINTICLGQAASMGSLLLCAGAKGQRRALPNA 144
D MQ + + ++T+C+G AASMGSLLL G +R A P+A
Sbjct: 313 DIMQVVEAEVHTVCVGLAASMGSLLLVGGEFTKRLAFPHA 432
>BQ139568 similar to SP|P12210|CLPP_ ATP-dependent Clp protease proteolytic
subunit (EC 3.4.21.92) (Endopeptidase Clp). [Common
tobacco], partial (41%)
Length = 714
Score = 68.9 bits (167), Expect = 2e-12
Identities = 28/72 (38%), Positives = 53/72 (72%)
Frame = -3
Query: 62 ISDDTAHVVVAQLLFLESENPSKPINMYLNSPGGAVTAGLAIYDTMQYIRSPINTICLGQ 121
++ + ++ +V +++L E+ ++ + +++NSPGG V +G+AI+D MQ + + ++T+C+G
Sbjct: 580 VNTEISNQLVGLMVYLSLEDKTRDLYLFINSPGGEVISGMAIFDIMQVVEAEVHTVCVGL 401
Query: 122 AASMGSLLLCAG 133
AASMGSLLL G
Sbjct: 400 AASMGSLLLVGG 365
>TC91885 similar to SP|Q9BBQ9|CLPP_LOTJA ATP-dependent Clp protease
proteolytic subunit (EC 3.4.21.92) (Endopeptidase Clp).
{Lotus japonicus}, partial (37%)
Length = 689
Score = 58.2 bits (139), Expect = 3e-09
Identities = 26/71 (36%), Positives = 46/71 (64%), Gaps = 1/71 (1%)
Frame = +2
Query: 146 IMIHQPSGG-YSGQAKDIAIHTKQIVRMWDALNELYVKHTGQPLDVIQKNMDRDYFMTAE 204
+MIHQP+ Y GQ + + +++++M + +Y + TG+ I ++M+RD FM+AE
Sbjct: 464 VMIHQPATSLYEGQVGECMLEAEELLKMRKTITNIYAQRTGKASWQIYRDMERDLFMSAE 643
Query: 205 EAKEFGIIDEV 215
EA+ +GI+D V
Sbjct: 644 EAEAYGIVDTV 676
>BQ141735 similar to PIR|S22697|S226 extensin - Volvox carteri (fragment),
partial (6%)
Length = 1127
Score = 26.6 bits (57), Expect = 9.1
Identities = 11/19 (57%), Positives = 14/19 (72%)
Frame = +2
Query: 136 GQRRALPNATIMIHQPSGG 154
G+RRA PN+TI I P+ G
Sbjct: 320 GRRRARPNSTIYITSPANG 376
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,108,166
Number of Sequences: 36976
Number of extensions: 74505
Number of successful extensions: 315
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 309
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 311
length of query: 238
length of database: 9,014,727
effective HSP length: 93
effective length of query: 145
effective length of database: 5,575,959
effective search space: 808514055
effective search space used: 808514055
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146777.12


