

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
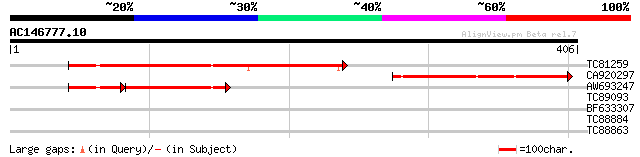
Query= AC146777.10 - phase: 1 /pseudo
(406 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC81259 weakly similar to PIR|T50512|T50512 hypothetical protein... 226 2e-59
CA920297 similar to GP|10177809|db gene_id:MKD15.6~unknown prote... 195 2e-50
AW693247 similar to GP|10177809|dbj gene_id:MKD15.6~unknown prot... 83 2e-24
TC89093 similar to GP|15011837|gb|AAK26164.2 calcium-dependent c... 29 2.9
BF633307 29 2.9
TC88884 29 2.9
TC88863 similar to GP|13161397|dbj|BAB33033. CPRD2 {Vigna unguic... 28 8.3
>TC81259 weakly similar to PIR|T50512|T50512 hypothetical protein T22D6.210
- Arabidopsis thaliana (fragment), partial (47%)
Length = 696
Score = 226 bits (575), Expect = 2e-59
Identities = 124/216 (57%), Positives = 153/216 (70%), Gaps = 16/216 (7%)
Frame = +3
Query: 43 SWFKCCFQAVKTLAKSPTFAFARDPRQLQYQIDLNRLFLYTSYNRLGKNASEADAEEIIE 102
S+F+ C +AV+TL + P+ +DPRQLQ++ D+NRLFLY+SYNRLGK+A E+D E+IIE
Sbjct: 57 SFFRKCREAVQTLGRRPSLT--KDPRQLQFEADVNRLFLYSSYNRLGKSADESDVEKIIE 230
Query: 103 IATKASVADQQMVVQENVHAQIKAFCTFMDAVFLPNEKKVNDVSFELSQQTKILPQHSDL 162
A+K S DQ VQENVH+QIK F MD + LPN+K VND LSQQ LP+ S L
Sbjct: 231 EASKTSFTDQVTQVQENVHSQIKTFSASMDEILLPNDKMVND-PLGLSQQESTLPRRSGL 407
Query: 163 RSANGKF-------------VIHQLTI-LKLKDELGYTLNVKPSQISHKDAGQGLFLDGV 208
A G+ + Q + KLKD+LG+TL+VKPSQISHKDAG+GLFLDGV
Sbjct: 408 SLAVGRTGSSPNNSAGPQTRPLSQAEVSQKLKDQLGFTLDVKPSQISHKDAGRGLFLDGV 587
Query: 209 VDVGAVVAFYPGVVYSPAYYHHIPGY--LDEQNPYL 242
DVG+VVAFYPGVVYSPAYY +IPGY + QNPYL
Sbjct: 588 ADVGSVVAFYPGVVYSPAYYRYIPGYPKVAAQNPYL 695
>CA920297 similar to GP|10177809|db gene_id:MKD15.6~unknown protein
{Arabidopsis thaliana}, partial (31%)
Length = 680
Score = 195 bits (496), Expect = 2e-50
Identities = 97/129 (75%), Positives = 111/129 (85%)
Frame = -2
Query: 275 KGSQVDNSDDVLERRNPLALAHFANHPSKGMLPNVMICPYDFPLIENDMRAYIPNVLFGN 334
+G Q D S +V+ERRNPLALAHFANHPSKG+LPNVMICPYDFPL EN++R Y+PN++FG+
Sbjct: 589 EGKQGD-SGEVIERRNPLALAHFANHPSKGVLPNVMICPYDFPLTENNIRVYVPNIMFGD 413
Query: 335 AAEENTERFGSFWFKSRVPRNNESHVPTTLKTVVLVATRALQDEELLLNYRLGNTKRCPE 394
AE RFGSFWFKS V +NN S VP TLK++VLVATRA+QDEELLLNYRL NTKR PE
Sbjct: 412 -AEVKMRRFGSFWFKSGVSKNNMSDVP-TLKSLVLVATRAIQDEELLLNYRLSNTKRRPE 239
Query: 395 WYAPVDEEE 403
WYAPVDEEE
Sbjct: 238 WYAPVDEEE 212
>AW693247 similar to GP|10177809|dbj gene_id:MKD15.6~unknown protein
{Arabidopsis thaliana}, partial (4%)
Length = 560
Score = 83.2 bits (204), Expect(2) = 2e-24
Identities = 44/75 (58%), Positives = 52/75 (68%)
Frame = +3
Query: 84 SYNRLGKNASEADAEEIIEIATKASVADQQMVVQENVHAQIKAFCTFMDAVFLPNEKKVN 143
SYNRLGK+A E+D E+IIE A+K S DQ VQENVH+QIK F MD + LPN+K VN
Sbjct: 174 SYNRLGKSADESDVEKIIEEASKTSFTDQVTQVQENVHSQIKTFSASMDEILLPNDKMVN 353
Query: 144 DVSFELSQQTKILPQ 158
D LSQQ LP+
Sbjct: 354 D-PLGLSQQESTLPR 395
Score = 47.0 bits (110), Expect(2) = 2e-24
Identities = 21/41 (51%), Positives = 32/41 (77%)
Frame = +1
Query: 43 SWFKCCFQAVKTLAKSPTFAFARDPRQLQYQIDLNRLFLYT 83
S+F+ C +AV+TL + P+ +DPRQLQ++ D+NRLFLY+
Sbjct: 43 SFFRKCREAVQTLGRRPSLT--KDPRQLQFEADVNRLFLYS 159
>TC89093 similar to GP|15011837|gb|AAK26164.2 calcium-dependent
calmodulin-independent protein kinase 5 {Cucumis
sativus}, partial (42%)
Length = 690
Score = 29.3 bits (64), Expect = 2.9
Identities = 12/20 (60%), Positives = 15/20 (75%)
Frame = +2
Query: 127 FCTFMDAVFLPNEKKVNDVS 146
FCTF VFLP+ +K+N VS
Sbjct: 191 FCTFSSLVFLPSGQKMNKVS 250
>BF633307
Length = 299
Score = 29.3 bits (64), Expect = 2.9
Identities = 19/65 (29%), Positives = 35/65 (53%), Gaps = 4/65 (6%)
Frame = -1
Query: 87 RLGKNASEADAEEIIEIATKASVADQQ----MVVQENVHAQIKAFCTFMDAVFLPNEKKV 142
R G NAS A + T A V++++ +V+Q+N H ++KA + + + K+
Sbjct: 224 RTGTNASYAARQ------TSACVSEKE*GDLVVLQKNCHMKVKAELSKASFLICLHHNKI 63
Query: 143 NDVSF 147
ND++F
Sbjct: 62 NDMAF 48
>TC88884
Length = 1128
Score = 29.3 bits (64), Expect = 2.9
Identities = 14/33 (42%), Positives = 20/33 (60%)
Frame = +2
Query: 13 LMFYHFSLALCEPKV*FLYLLCIL*SLLQKSWF 45
L +HF +ALCE KV + +L C L ++ WF
Sbjct: 785 LFSFHFLVALCEGKVPY-HLFCFLQKKMEGFWF 880
>TC88863 similar to GP|13161397|dbj|BAB33033. CPRD2 {Vigna unguiculata},
partial (33%)
Length = 698
Score = 27.7 bits (60), Expect = 8.3
Identities = 12/42 (28%), Positives = 22/42 (51%), Gaps = 3/42 (7%)
Frame = +2
Query: 286 LERRNPLALAHFANHP---SKGMLPNVMICPYDFPLIENDMR 324
++ +NPL+L+H P +PN M C ++F ++ R
Sbjct: 233 MKHQNPLSLSHLHKFPIFKQP*SVPNTMACKFEFVVVATTSR 358
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.324 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,318,486
Number of Sequences: 36976
Number of extensions: 191554
Number of successful extensions: 847
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 837
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 844
length of query: 406
length of database: 9,014,727
effective HSP length: 98
effective length of query: 308
effective length of database: 5,391,079
effective search space: 1660452332
effective search space used: 1660452332
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146777.10


