

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146776.7 + phase: 0
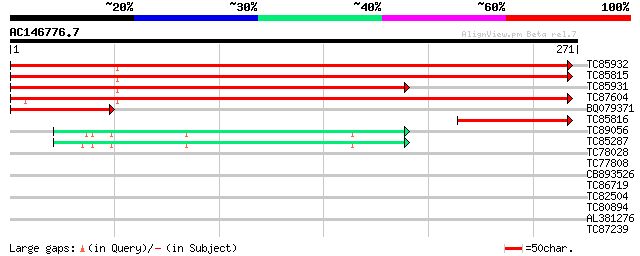
(271 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85932 homologue to GP|10177452|dbj|BAB10843. hypersensitive-in... 481 e-136
TC85815 homologue to GP|3928150|emb|CAA10289.1 hypothetical prot... 428 e-120
TC85931 similar to GP|10177452|dbj|BAB10843. hypersensitive-indu... 350 3e-97
TC87604 similar to GP|9758199|dbj|BAB08673.1 gb|AAF03497.1~gene_... 290 5e-79
BQ079371 similar to GP|7716470|gb|A hypersensitive-induced respo... 103 5e-23
TC85816 similar to GP|15021744|gb|AAK77899.1 root nodule extensi... 84 4e-17
TC89056 similar to GP|12751303|gb|AAK07610.1 prohibitin 1-like p... 41 4e-04
TC85287 similar to SP|O50008|METE_ARATH 5-methyltetrahydropteroy... 40 7e-04
TC78028 similar to GP|22136524|gb|AAM91340.1 unknown protein {Ar... 31 0.44
TC77808 similar to PIR|T03843|T03843 prohibitin - common tobacco... 30 1.3
CB893526 similar to GP|12751303|gb| prohibitin 1-like protein {B... 28 3.7
TC86719 similar to GP|599956|emb|CAA83434.1| chloroplast inner m... 28 3.7
TC82504 similar to GP|13899105|gb|AAK48974.1 putative protein {A... 28 4.9
TC80894 weakly similar to GP|18855010|gb|AAL79702.1 unknown prot... 28 4.9
AL381276 similar to GP|2582388|gb| prohibitin {Pneumocystis cari... 27 6.4
TC87239 similar to PIR|T09028|T09028 hypothetical protein T27E11... 27 8.3
>TC85932 homologue to GP|10177452|dbj|BAB10843. hypersensitive-induced
response protein {Arabidopsis thaliana}, partial (98%)
Length = 1555
Score = 481 bits (1238), Expect = e-136
Identities = 250/282 (88%), Positives = 257/282 (90%), Gaps = 13/282 (4%)
Frame = +2
Query: 1 MGNIVCCVQVDQSQVAMKEGFGKFEKVLHPGCHCMPWFLGKRIAGHLSLR---------- 50
MGN++CCVQVDQS VAMKEGFGKFE+VL PGCHCMPWFLGKRIAGHLSLR
Sbjct: 269 MGNLLCCVQVDQSTVAMKEGFGKFEEVLQPGCHCMPWFLGKRIAGHLSLRLQQLDIKCET 448
Query: 51 ---DNVFVNVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQ 107
DNVFVNVVASIQYRALA+ ANDAFYKLSNTRGQIQAYVFDVIRASVPKL LDD FEQ
Sbjct: 449 KTKDNVFVNVVASIQYRALADNANDAFYKLSNTRGQIQAYVFDVIRASVPKLYLDDAFEQ 628
Query: 108 KNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEA 167
KNEIAKAVEEE EKAMSAYGYEIVQTLITDIEPD HVK AMNEINAAARMR+AA EKAEA
Sbjct: 629 KNEIAKAVEEELEKAMSAYGYEIVQTLITDIEPDDHVKRAMNEINAAARMRLAAKEKAEA 808
Query: 168 EKILQIKRAEGEAESKYLSGMGIARQRQAIVDGLRDSVIGFSENVPGPSAKDVMDMVLVT 227
EKILQIKRAEGEAESKYLSG+GIARQRQAIVDGLRDSVIGFS NVPG +AKDVMDMVLVT
Sbjct: 809 EKILQIKRAEGEAESKYLSGLGIARQRQAIVDGLRDSVIGFSVNVPGTTAKDVMDMVLVT 988
Query: 228 QYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQICDGLLQGS 269
QYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQI DGLLQGS
Sbjct: 989 QYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQIRDGLLQGS 1114
>TC85815 homologue to GP|3928150|emb|CAA10289.1 hypothetical protein {Cicer
arietinum}, complete
Length = 1245
Score = 428 bits (1100), Expect = e-120
Identities = 222/282 (78%), Positives = 245/282 (86%), Gaps = 13/282 (4%)
Frame = +1
Query: 1 MGNIVCCVQVDQSQVAMKEGFGKFEKVLHPGCHCMPWFLGKRIAGHLSLR---------- 50
MG + CVQVDQS VA+KE FGKF VL PGCHC+PW LG +IAG LSLR
Sbjct: 133 MGQALGCVQVDQSNVAIKEHFGKFADVLEPGCHCLPWCLGYQIAGGLSLRVQQLDVKCET 312
Query: 51 ---DNVFVNVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQ 107
DNVFVNVVAS+QYRA+A+KA+DAFY+L+NTR QIQ+YVFDVIRASVPKL LD FEQ
Sbjct: 313 KTKDNVFVNVVASVQYRAVADKASDAFYRLTNTREQIQSYVFDVIRASVPKLELDAVFEQ 492
Query: 108 KNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEA 167
KN+IAKAVEEE EKAMS YGY+IVQTLI DIEPD +VK AMNEINAAARMR+AANEKAEA
Sbjct: 493 KNDIAKAVEEELEKAMSMYGYQIVQTLIVDIEPDVNVKRAMNEINAAARMRLAANEKAEA 672
Query: 168 EKILQIKRAEGEAESKYLSGMGIARQRQAIVDGLRDSVIGFSENVPGPSAKDVMDMVLVT 227
EKILQIK+AEGEAESKYLSG+GIARQRQAIVDGLRDSV+ FSENVPG +AKDVMDMVLVT
Sbjct: 673 EKILQIKKAEGEAESKYLSGLGIARQRQAIVDGLRDSVLAFSENVPGTTAKDVMDMVLVT 852
Query: 228 QYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQICDGLLQGS 269
QYFDTMKEIGA+SKSS+VFIPHGPGAVRD+A QI DGLLQG+
Sbjct: 853 QYFDTMKEIGASSKSSSVFIPHGPGAVRDIAVQIRDGLLQGN 978
>TC85931 similar to GP|10177452|dbj|BAB10843. hypersensitive-induced
response protein {Arabidopsis thaliana}, partial (71%)
Length = 779
Score = 350 bits (898), Expect = 3e-97
Identities = 181/204 (88%), Positives = 185/204 (89%), Gaps = 13/204 (6%)
Frame = +1
Query: 1 MGNIVCCVQVDQSQVAMKEGFGKFEKVLHPGCHCMPWFLGKRIAGHLSLR---------- 50
MGN+VCCVQVDQSQVAMKEGFGKFEKVL PGCHCMPWFLGKRIAGHLSLR
Sbjct: 166 MGNLVCCVQVDQSQVAMKEGFGKFEKVLQPGCHCMPWFLGKRIAGHLSLRVQQLDIKCET 345
Query: 51 ---DNVFVNVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTFEQ 107
DNVFVNVVASIQYRALA+KANDAFYKLSNTR QIQAYVFDVIRASVPKLNLDDTFEQ
Sbjct: 346 KTKDNVFVNVVASIQYRALADKANDAFYKLSNTRNQIQAYVFDVIRASVPKLNLDDTFEQ 525
Query: 108 KNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEA 167
KNEIAKAVEEE EKAMSAYGYEIVQTLITDIEPD HVK AMNEINAAARMR+AA EKAEA
Sbjct: 526 KNEIAKAVEEELEKAMSAYGYEIVQTLITDIEPDVHVKRAMNEINAAARMRLAAKEKAEA 705
Query: 168 EKILQIKRAEGEAESKYLSGMGIA 191
EKILQIKRAEGEAESKYLSG+GIA
Sbjct: 706 EKILQIKRAEGEAESKYLSGLGIA 777
>TC87604 similar to GP|9758199|dbj|BAB08673.1
gb|AAF03497.1~gene_id:K17N15.12~similar to unknown
protein {Arabidopsis thaliana}, partial (96%)
Length = 1194
Score = 290 bits (741), Expect = 5e-79
Identities = 149/284 (52%), Positives = 198/284 (69%), Gaps = 15/284 (5%)
Frame = +3
Query: 1 MGNIVC--CVQVDQSQVAMKEGFGKFEKVLHPGCHCMPWFLGKRIAGHLSLR-------- 50
MGN C C V+QS V + E +G+F++V PG F G+ +AG LS R
Sbjct: 129 MGNTFCLFCGCVEQSSVGIVEQWGRFQRVAQPGFQIFNPFAGECLAGILSTRIASLDVKI 308
Query: 51 -----DNVFVNVVASIQYRALANKANDAFYKLSNTRGQIQAYVFDVIRASVPKLNLDDTF 105
DNVFV ++ SIQYR + A+DAFY+L N + QIQAYVFDV RA VPK+NLD+ F
Sbjct: 309 ETKTKDNVFVQLLCSIQYRVVKENADDAFYELQNPQEQIQAYVFDVARAIVPKMNLDELF 488
Query: 106 EQKNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKA 165
EQK E+AK V EE K M YGY I L+ DI PD V+ AMNEINAA R+ +A+ K
Sbjct: 489 EQKGEVAKGVMEELGKVMGEYGYSIEHILMVDIIPDPSVRRAMNEINAAQRLLLASEFKG 668
Query: 166 EAEKILQIKRAEGEAESKYLSGMGIARQRQAIVDGLRDSVIGFSENVPGPSAKDVMDMVL 225
EA+K+L +K+AE EAESK+L G+G+ARQRQAI DGLR++++ FS V G SAK+VMD+++
Sbjct: 669 EADKVLIVKKAEAEAESKFLGGVGVARQRQAITDGLRENILQFSNKVEGTSAKEVMDLIM 848
Query: 226 VTQYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQICDGLLQGS 269
+TQYFDT++++G SK++ VFIPHGPG VRD+ QI +G+++ S
Sbjct: 849 ITQYFDTIRDLGNNSKNTTVFIPHGPGHVRDIGDQIRNGMMEAS 980
>BQ079371 similar to GP|7716470|gb|A hypersensitive-induced response protein
{Zea mays}, partial (21%)
Length = 397
Score = 103 bits (258), Expect = 5e-23
Identities = 45/50 (90%), Positives = 48/50 (96%)
Frame = +2
Query: 1 MGNIVCCVQVDQSQVAMKEGFGKFEKVLHPGCHCMPWFLGKRIAGHLSLR 50
MGN++CCVQVDQS VAMKEGFGKFE+VL PGCHCMPWFLGKRIAGHLSLR
Sbjct: 77 MGNLLCCVQVDQSTVAMKEGFGKFEEVLQPGCHCMPWFLGKRIAGHLSLR 226
>TC85816 similar to GP|15021744|gb|AAK77899.1 root nodule extensin {Pisum
sativum}, partial (32%)
Length = 603
Score = 84.3 bits (207), Expect = 4e-17
Identities = 41/55 (74%), Positives = 46/55 (83%)
Frame = +2
Query: 215 PSAKDVMDMVLVTQYFDTMKEIGAASKSSAVFIPHGPGAVRDVASQICDGLLQGS 269
P + VLVTQYFDTMKEIGA+SKSS+VFIPHGPGAVRD+A QI DGLLQG+
Sbjct: 173 PLSTTTSSQVLVTQYFDTMKEIGASSKSSSVFIPHGPGAVRDIAVQIRDGLLQGN 337
>TC89056 similar to GP|12751303|gb|AAK07610.1 prohibitin 1-like protein
{Brassica napus}, partial (96%)
Length = 1131
Score = 41.2 bits (95), Expect = 4e-04
Identities = 43/186 (23%), Positives = 74/186 (39%), Gaps = 16/186 (8%)
Frame = +1
Query: 22 GKFEKVLHPGCHCM-PWF-----LGKRIAGHL-----SLRDNVFVNVVASIQYRALANKA 70
G +KV G H M PWF R HL RD V + + R + ++
Sbjct: 244 GVKDKVYPEGTHIMIPWFERPVIYDVRARPHLVESTSGSRDLQMVKIGLRVLTRPVPDQL 423
Query: 71 NDAFYKLSNTRGQ--IQAYVFDVIRASVPKLNLDDTFEQKNEIAKAVEEEREKAMSAYGY 128
+ L + + + + + +++ V + N Q+ +++ + + + S +
Sbjct: 424 PTVYRTLGENYNERVLPSIIHETLKSVVAQYNASQLITQREAVSREIRKILTERASQFNI 603
Query: 129 EIVQTLITDIEPDQHVKTAMNEINAAARMRIAAN---EKAEAEKILQIKRAEGEAESKYL 185
+ IT + + A+ AA+ A EKAE +K I RA+GEA S L
Sbjct: 604 ALDDVSITSLTFGREFTAAIEAKQVAAQEAERAKFVVEKAEQDKRSAIIRAQGEATSAQL 783
Query: 186 SGMGIA 191
G IA
Sbjct: 784 IGQAIA 801
>TC85287 similar to SP|O50008|METE_ARATH
5-methyltetrahydropteroyltriglutamate--homocysteine
methyltransferase (EC 2.1.1.14), complete
Length = 3967
Score = 40.4 bits (93), Expect = 7e-04
Identities = 42/186 (22%), Positives = 74/186 (39%), Gaps = 16/186 (8%)
Frame = +3
Query: 22 GKFEKVLHPGCH-CMPWF-----LGKRIAGHL-----SLRDNVFVNVVASIQYRALANKA 70
G +KV G H +PWF R HL RD V + + R L +
Sbjct: 246 GVKDKVYPEGTHFVIPWFERPVIYDVRARPHLVESTSGSRDLQMVKIGLRVLTRPLPGQL 425
Query: 71 NDAFYKLSNTRGQ--IQAYVFDVIRASVPKLNLDDTFEQKNEIAKAVEEEREKAMSAYGY 128
+ L + + + + + ++A V + N Q+ +++ + + + + +
Sbjct: 426 PTVYRTLGENYNERVLPSIIHETLKAVVAQYNASQLITQREAVSREIRKILTERAANFNI 605
Query: 129 EIVQTLITDIEPDQHVKTAMNEINAAARMRIAAN---EKAEAEKILQIKRAEGEAESKYL 185
+ IT + + A+ AA+ A EKAE +K + RA+GEA+S L
Sbjct: 606 ALDDVSITSLTFGKEFTAAIEAKQVAAQEAERAKFVVEKAEQDKRSAVIRAQGEAKSAQL 785
Query: 186 SGMGIA 191
G IA
Sbjct: 786 IGQAIA 803
>TC78028 similar to GP|22136524|gb|AAM91340.1 unknown protein {Arabidopsis
thaliana}, partial (86%)
Length = 1217
Score = 31.2 bits (69), Expect = 0.44
Identities = 22/91 (24%), Positives = 46/91 (50%)
Frame = +1
Query: 130 IVQTLITDIEPDQHVKTAMNEINAAARMRIAANEKAEAEKILQIKRAEGEAESKYLSGMG 189
+ + +TD+E D + A +E+ +A +A N E++ +E + SGM
Sbjct: 160 VYKGFLTDLECDHLISIAKSELKRSA---VADNLSGESKL----------SEVRTSSGMF 300
Query: 190 IARQRQAIVDGLRDSVIGFSENVPGPSAKDV 220
I++ + AIV G+ D + ++ +P + +D+
Sbjct: 301 ISKNKDAIVSGIEDKISSWT-FLPKENGEDI 390
>TC77808 similar to PIR|T03843|T03843 prohibitin - common tobacco, partial
(96%)
Length = 1202
Score = 29.6 bits (65), Expect = 1.3
Identities = 25/116 (21%), Positives = 49/116 (41%), Gaps = 10/116 (8%)
Frame = +3
Query: 90 DVIRASVPKLNLDDTFEQKNEIAKAVEEEREKAMSAYGYEIVQTLITDIEPDQHVKTAMN 149
+V++A V + N D + +++ V + + + + IT + A+
Sbjct: 456 EVLKAVVAQFNADQLLTDRPQVSALVRDSLVRRAKDFNILLDDVAITHLSYGGEFSRAVE 635
Query: 150 EINAAARMRIAAN---EKAEAEKILQIKRAEGEAESKYL-------SGMGIARQRQ 195
+ A + + KAE E+ I RAEGE+++ L +GMG+ R+
Sbjct: 636 QKQVAQQEAERSKFVVMKAEQERRAAIIRAEGESDAAKLISDATAVAGMGLIELRR 803
>CB893526 similar to GP|12751303|gb| prohibitin 1-like protein {Brassica
napus}, partial (14%)
Length = 127
Score = 28.1 bits (61), Expect = 3.7
Identities = 14/25 (56%), Positives = 17/25 (68%)
Frame = +2
Query: 163 EKAEAEKILQIKRAEGEAESKYLSG 187
EKAE EK + RA+GEA+S L G
Sbjct: 47 EKAEQEKRSAVTRAQGEAKSAQLIG 121
>TC86719 similar to GP|599956|emb|CAA83434.1| chloroplast inner membrane
protein {Pisum sativum}, partial (63%)
Length = 2950
Score = 28.1 bits (61), Expect = 3.7
Identities = 18/54 (33%), Positives = 30/54 (55%), Gaps = 3/54 (5%)
Frame = +3
Query: 77 LSNTRGQIQAYVFDVIRASV--PKLNLDDTFEQKNE-IAKAVEEEREKAMSAYG 127
+S QI+ ++++S PKLNLD T ++ E + K V++E +A A G
Sbjct: 1572 VSEVEVQIEKLKQQILKSSSQPPKLNLDKTIKKLEEKLEKEVDQELSEAAKALG 1733
>TC82504 similar to GP|13899105|gb|AAK48974.1 putative protein {Arabidopsis
thaliana}, partial (59%)
Length = 714
Score = 27.7 bits (60), Expect = 4.9
Identities = 23/66 (34%), Positives = 35/66 (52%), Gaps = 1/66 (1%)
Frame = +3
Query: 156 RMRIAANEKAEAEKILQIKRAEGEAESKYLSGMGIARQRQAIVDGLRDSVIGFSENVP-G 214
R + + NE+ EA K +IKR + E E +G+A +R + G R FSE V G
Sbjct: 270 RDKKSQNEEMEAAKE-EIKRIKEEEEQAMREALGLAPKRASRPQGNRLDKHEFSELVKRG 446
Query: 215 PSAKDV 220
+A+D+
Sbjct: 447 STAEDL 464
>TC80894 weakly similar to GP|18855010|gb|AAL79702.1 unknown protein {Oryza
sativa}, partial (20%)
Length = 679
Score = 27.7 bits (60), Expect = 4.9
Identities = 22/69 (31%), Positives = 35/69 (49%), Gaps = 2/69 (2%)
Frame = +3
Query: 120 EKAMSAYGYEIVQTLITDIEP--DQHVKTAMNEINAAARMRIAANEKAEAEKILQIKRAE 177
+KA+ +G E+ + E +Q + A EI R ++ ANE+ + E IK E
Sbjct: 75 KKALDEHGLELRVRAANEAEAACEQRLSAAEAEIEEL-RAQLDANERKKLEMTEAIKAKE 251
Query: 178 GEAESKYLS 186
EAE+ Y+S
Sbjct: 252 AEAET-YIS 275
>AL381276 similar to GP|2582388|gb| prohibitin {Pneumocystis carinii},
partial (37%)
Length = 449
Score = 27.3 bits (59), Expect = 6.4
Identities = 14/20 (70%), Positives = 15/20 (75%)
Frame = +2
Query: 163 EKAEAEKILQIKRAEGEAES 182
EKAE EK I RAEGEAE+
Sbjct: 125 EKAEQEKQAAIIRAEGEAEA 184
>TC87239 similar to PIR|T09028|T09028 hypothetical protein T27E11.130 -
Arabidopsis thaliana, partial (69%)
Length = 1228
Score = 26.9 bits (58), Expect = 8.3
Identities = 14/38 (36%), Positives = 22/38 (57%)
Frame = +1
Query: 144 VKTAMNEINAAARMRIAANEKAEAEKILQIKRAEGEAE 181
VK + AA + +IAA EKA+ +K + AE +A+
Sbjct: 271 VKVKKAKAVAAEKAKIAAEEKAKVDKAASVAAAEKKAK 384
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.133 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,088,988
Number of Sequences: 36976
Number of extensions: 62167
Number of successful extensions: 369
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 365
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 365
length of query: 271
length of database: 9,014,727
effective HSP length: 95
effective length of query: 176
effective length of database: 5,502,007
effective search space: 968353232
effective search space used: 968353232
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146776.7


