

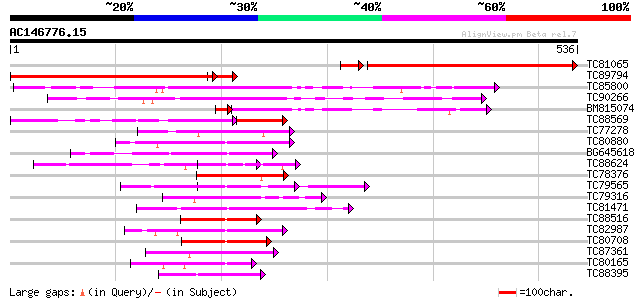
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146776.15 + phase: 0
(536 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC81065 weakly similar to GP|11320830|dbj|BAB18313. putative WRK... 396 e-117
TC89794 similar to PIR|T49948|T49948 hypothetical protein F8M21.... 378 e-116
TC85800 similar to GP|11320830|dbj|BAB18313. putative WRKY DNA b... 230 1e-60
TC90266 similar to SP|Q9C519|WRK6_ARATH WRKY transcription facto... 160 1e-39
BM815074 similar to GP|13506739|gb WRKY DNA-binding protein 6 {A... 110 4e-29
TC88569 homologue to SP|Q9C519|WRK6_ARATH WRKY transcription fac... 109 3e-24
TC77278 similar to GP|11493822|gb|AAG35658.1 transcription facto... 105 4e-23
TC80880 similar to SP|Q9ZQ70|WRK3_ARATH Probable WRKY transcript... 105 6e-23
BG645618 similar to GP|18158619|gb WRKY-like drought-induced pro... 103 1e-22
TC88624 similar to PIR|S51529|S51529 SPF1 protein - sweet potato... 101 8e-22
TC78376 similar to SP|Q9SK33|WR60_ARATH Probable WRKY transcript... 100 2e-21
TC79565 similar to SP|Q9ZQ70|WRK3_ARATH Probable WRKY transcript... 96 3e-20
TC79316 similar to GP|11493822|gb|AAG35658.1 transcription facto... 95 6e-20
TC81471 similar to PIR|A84913|A84913 probable WRKY-type DNA bind... 95 6e-20
TC88516 similar to GP|14530681|dbj|BAB61053. WRKY DNA-binding pr... 94 2e-19
TC82987 similar to GP|15991740|gb|AAL13047.1 WRKY transcription ... 93 2e-19
TC80708 weakly similar to GP|14530685|dbj|BAB61055. WRKY DNA-bin... 89 6e-18
TC87361 similar to GP|15991736|gb|AAL13045.1 WRKY transcription ... 89 6e-18
TC80165 similar to PIR|A84913|A84913 probable WRKY-type DNA bind... 86 4e-17
TC88395 similar to SP|Q9LP56|WR65_ARATH Probable WRKY transcript... 85 8e-17
>TC81065 weakly similar to GP|11320830|dbj|BAB18313. putative WRKY DNA
binding protein {Oryza sativa (japonica
cultivar-group)}, partial (4%)
Length = 828
Score = 396 bits (1017), Expect(2) = e-117
Identities = 198/198 (100%), Positives = 198/198 (100%)
Frame = +1
Query: 339 PRYPSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLNFGRQNTMESIYQT 398
PRYPSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLNFGRQNTMESIYQT
Sbjct: 82 PRYPSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLNFGRQNTMESIYQT 261
Query: 399 YMQKNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAAALSSLIGNTTNQGNQNQSA 458
YMQKNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAAALSSLIGNTTNQGNQNQSA
Sbjct: 262 YMQKNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAAALSSLIGNTTNQGNQNQSA 441
Query: 459 GENLSQKMKWAEMFQVSSTSLPSSSSKVNGCASSFLNKTAPVNNTQNGSLMLLSPSLPFS 518
GENLSQKMKWAEMFQVSSTSLPSSSSKVNGCASSFLNKTAPVNNTQNGSLMLLSPSLPFS
Sbjct: 442 GENLSQKMKWAEMFQVSSTSLPSSSSKVNGCASSFLNKTAPVNNTQNGSLMLLSPSLPFS 621
Query: 519 ATKSASTSPGGDNSDNTN 536
ATKSASTSPGGDNSDNTN
Sbjct: 622 ATKSASTSPGGDNSDNTN 675
Score = 44.3 bits (103), Expect(2) = e-117
Identities = 20/22 (90%), Positives = 20/22 (90%)
Frame = +2
Query: 313 TSNPSNSSTSSPFVRFNSSYNN 334
TSNPSNSSTSSP VRFN SYNN
Sbjct: 2 TSNPSNSSTSSPLVRFNPSYNN 67
>TC89794 similar to PIR|T49948|T49948 hypothetical protein F8M21.20 -
Arabidopsis thaliana, partial (12%)
Length = 892
Score = 378 bits (970), Expect(2) = e-116
Identities = 186/197 (94%), Positives = 192/197 (97%), Gaps = 1/197 (0%)
Frame = +3
Query: 1 MGEVREENQRLKMCLNKIMTEYRALEMQFNNMVKQETKKNNDNNHEEMNAESDLVSLSLG 60
MGEVREENQRLKMCLNKIMTEYRALEMQFNNMVKQETKKNNDNNHEEMNAESDLVSLSLG
Sbjct: 246 MGEVREENQRLKMCLNKIMTEYRALEMQFNNMVKQETKKNNDNNHEEMNAESDLVSLSLG 425
Query: 61 RVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNIPSP 120
RVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNIPSP
Sbjct: 426 RVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNIPSP 605
Query: 121 VNSSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTMNDG 180
VNSSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTMNDG
Sbjct: 606 VNSSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTMNDG 785
Query: 181 CQWRK-YGQKIAKGNPC 196
CQW++ + +K+ + PC
Sbjct: 786 CQWKEVWTKKLQREIPC 836
Score = 60.1 bits (144), Expect(2) = e-116
Identities = 26/28 (92%), Positives = 27/28 (95%)
Frame = +2
Query: 188 QKIAKGNPCPRAYYRCTVAPSCPVRKQV 215
+KIAKGNP PRAYYRCTVAPSCPVRKQV
Sbjct: 809 KKIAKGNPLPRAYYRCTVAPSCPVRKQV 892
>TC85800 similar to GP|11320830|dbj|BAB18313. putative WRKY DNA binding
protein {Oryza sativa (japonica cultivar-group)},
partial (37%)
Length = 2028
Score = 230 bits (586), Expect = 1e-60
Identities = 174/468 (37%), Positives = 236/468 (50%), Gaps = 8/468 (1%)
Frame = +1
Query: 4 VREENQRLKMCLNKIMTEYRALEMQFNNMVKQETKKNNDNNHEEMNAESDLVSLSLGRVP 63
+ EN++LK L+ + Y AL++Q +++ KN+ +E +NA+++ + +G
Sbjct: 568 MNSENKKLKEMLSHVTGNYTALQLQLVALMQ----KNHHTENEVVNAKAEEKNQGVGGAM 735
Query: 64 SNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNIPSPVNS 123
+P+ E N + E + + S S S N P NS
Sbjct: 736 ---VPRQFLEITNGTT-------------------EVEDQVSNSSSDERTRSNTPQMRNS 849
Query: 124 SEVVPIKNDEVVET--WPPSKT---LNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTMN 178
+ ++ ET W P+K+ LN + + D+ + +KARV VRAR + ++
Sbjct: 850 NGKTGREDSPESETQGWGPNKSQKILNSS--NVADQANTEATMRKARVSVRARSEASMIS 1023
Query: 179 DGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLP 238
DGCQWRKYGQK+AKGNPCPRAYYRCT+A CPVRKQVQRC ED +IL+TTYEGTHNH LP
Sbjct: 1024DGCQWRKYGQKMAKGNPCPRAYYRCTMAVGCPVRKQVQRCAEDKTILVTTYEGTHNHPLP 1203
Query: 239 LSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNFYLPDGTKSNQLYLSNPA 298
+A AMASTTSAAASMLLSGS +S G M T N L LP T S
Sbjct: 1204PAAMAMASTTSAAASMLLSGSMSSADGIM----TPNLL--ARAILPCST-------SMAT 1344
Query: 299 LSSQHSHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQLPRYPSSTLSFSSPESNPMHW 358
LS+ PT+TLDLT N S+P + L F P+ P H
Sbjct: 1345LSASAPFPTVTLDLTQN------SNP-------------------NPLQFQRPQHAPFHQ 1449
Query: 359 NSFLNYATTQN--QPYSNNRNNNNLSTLNFGRQNTMESIYQTYMQKNNNSSNISQHVGLQ 416
QN Q ++ N + S L ++ + S + T S+ Q L
Sbjct: 1450VPSFFQGQNQNFAQAAASLYNQSKFSGLQLSQE--VGSSHLT----TQASTQQQQQPSLA 1611
Query: 417 DSTISAATKAITADPTFQSALAAALSSLI-GNTTNQGNQNQSAGENLS 463
DS +SAAT AITADP F + LAAA+SS+I G N N N ++ +S
Sbjct: 1612DS-VSAATAAITADPNFTAVLAAAISSIIGGGHANNSNNNSNSRNTIS 1752
>TC90266 similar to SP|Q9C519|WRK6_ARATH WRKY transcription factor 6 (WRKY
DNA-binding protein 6) (AtWRKY6). [Mouse-ear cress],
partial (15%)
Length = 1376
Score = 160 bits (404), Expect = 1e-39
Identities = 129/425 (30%), Positives = 190/425 (44%), Gaps = 10/425 (2%)
Frame = +2
Query: 36 ETKKNNDNNHEEMNAESDLVSLSLGRVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELS 95
E+ K+N+N +N +D ++ R+ +++ S L+ NN KE
Sbjct: 47 ESYKDNENLRGMLNLVNDRCNVLQNRLLL-------AMHMHQSSSLSQNNHNLLLKENTQ 205
Query: 96 LGLECKFETSKSGSTTEGLPNIPSPVNSSE---VVPIKNDE------VVETWPPSKTLNK 146
+ KS T + PSP N S+ ++N+E + + +N
Sbjct: 206 -------DAGKSVLPTRQFFDEPSPSNCSKNNGFAIVENNENNMGRNLACEYINEGEINS 364
Query: 147 TMRDAEDEVAQQTPAKKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVA 206
+ D EV ++ARV +RAR D M DGCQWRKYGQK AKGNPCPRAYYRC++
Sbjct: 365 KIEDQSSEVG----CRRARVSIRARSDFAFMVDGCQWRKYGQKTAKGNPCPRAYYRCSMG 532
Query: 207 PSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGS 266
SCPVRKQVQRC +D S+ ITTYEG HNH LP +A +A+ TS+A + L SST+
Sbjct: 533 TSCPVRKQVQRCFKDESVFITTYEGNHNHQLPPAAKPIANLTSSALNTFLPTSSTT---- 700
Query: 267 MPSAQTNNNLHGLNFYLPDGTKSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFV 326
Q NNL +N S+P +P NS+ + F
Sbjct: 701 --LQQYGNNL------------TNTFLFSSPL----------------SPPNSNAIATF- 787
Query: 327 RFNSSYNNNNQLPRYPSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLNF 386
+ P P+ TL F+ P SN Y +N+ S L F
Sbjct: 788 ---------SPSPTCPTITLDFTLPPSN-----------------YLQFKNHKQSSLLPF 889
Query: 387 GRQNTMESIYQTYMQKNNNSSNISQHVGLQDSTISAATKAITADPTFQSALAAALSSLI- 445
Q ++ + NN + + + ++A+ DP+ + AL +A+SS
Sbjct: 890 PFQGHYNQSFEVFPNMINNERKL--------ALVDVVSEALEKDPSLKEALFSAMSSFTN 1045
Query: 446 GNTTN 450
G+++N
Sbjct: 1046GDSSN 1060
>BM815074 similar to GP|13506739|gb WRKY DNA-binding protein 6 {Arabidopsis
thaliana}, partial (62%)
Length = 687
Score = 110 bits (274), Expect(2) = 4e-29
Identities = 94/255 (36%), Positives = 120/255 (46%), Gaps = 9/255 (3%)
Frame = +2
Query: 210 PVRKQVQRCVEDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPS 269
PVRKQVQRC ED SILITTYEGTH+H LP +A MASTT+AAA++LLSGS +S G M
Sbjct: 50 PVRKQVQRCAEDRSILITTYEGTHSHPLPPAAMPMASTTAAAATVLLSGSMSSADGVM-- 223
Query: 270 AQTNNNLHGLNFYLPDGTKSNQLYLSNPALSSQHSHPTITLDLTSNPSNSSTSSPFVRFN 329
N NL L LP+ + S LS+ PT+TLDLT + ++++ +SP
Sbjct: 224 ---NPNL--LARILPNCSS------SMATLSASAPFPTVTLDLTRDTTDNNGNSP----- 355
Query: 330 SSYNNNNQLPRYPSSTLSFSSPESNPMHWNSFLNYATTQNQPYSNNRNNNNLSTLNFGRQ 389
S P+ NFG
Sbjct: 356 --------------SQFQLGQPQ--------------------------------NFGSG 397
Query: 390 NTMESIYQT-YMQKNNNSSNISQHVG--------LQDSTISAATKAITADPTFQSALAAA 440
+ I Q Y Q + +SQ VG Q S++SA AITADP F +ALAAA
Sbjct: 398 QLPQVIAQALYNQSKFSGLQMSQDVGGSSQLHPTQQASSLSA---AITADPNFTAALAAA 568
Query: 441 LSSLIGNTTNQGNQN 455
+SS+IG N +
Sbjct: 569 ISSIIGAAPPSNNNS 613
Score = 36.6 bits (83), Expect(2) = 4e-29
Identities = 13/16 (81%), Positives = 14/16 (87%)
Frame = +1
Query: 195 PCPRAYYRCTVAPSCP 210
PCPRAYYRCT+A CP
Sbjct: 4 PCPRAYYRCTMALGCP 51
>TC88569 homologue to SP|Q9C519|WRK6_ARATH WRKY transcription factor 6 (WRKY
DNA-binding protein 6) (AtWRKY6). [Mouse-ear cress],
partial (21%)
Length = 1319
Score = 109 bits (272), Expect = 3e-24
Identities = 71/215 (33%), Positives = 106/215 (49%)
Frame = +2
Query: 1 MGEVREENQRLKMCLNKIMTEYRALEMQFNNMVKQETKKNNDNNHEEMNAESDLVSLSLG 60
+G V ENQ+LK L+ + + Y L +F ++++Q+ + +++H
Sbjct: 518 LGRVNAENQKLKDMLSDMNSSYTNLHNRFISLMQQQQNQTTEHDHI-------------- 655
Query: 61 RVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNIPSP 120
V + K D V++ +N E + T + P P
Sbjct: 656 -VNGKAVEKGD----GVVARKFMNGP----------AAEVDDQQEPEPCTPQNNHKEPDP 790
Query: 121 VNSSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTMNDG 180
++SE+V + + + PS +A D+ + +KARV VRAR + +NDG
Sbjct: 791 -DASELVQLLDRSQLPRLNPS--------NAADQANAEATMRKARVSVRARSEAHMINDG 943
Query: 181 CQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQV 215
CQWRKYGQK+AKGNPCPRAYYRCT+A CPVRKQV
Sbjct: 944 CQWRKYGQKMAKGNPCPRAYYRCTMALGCPVRKQV 1048
Score = 71.2 bits (173), Expect = 9e-13
Identities = 35/48 (72%), Positives = 41/48 (84%)
Frame = +1
Query: 215 VQRCVEDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTS 262
VQRC ED SILITTYEGTH+H LP +A MASTT+AAA++LLSGS +S
Sbjct: 1171 VQRCAEDRSILITTYEGTHSHPLPPAAMPMASTTAAAATVLLSGSMSS 1314
>TC77278 similar to GP|11493822|gb|AAG35658.1 transcription factor WRKY4
{Petroselinum crispum}, partial (40%)
Length = 1604
Score = 105 bits (262), Expect = 4e-23
Identities = 60/165 (36%), Positives = 85/165 (51%), Gaps = 17/165 (10%)
Frame = +2
Query: 122 NSSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTM--ND 179
N+S ++ I N + ++ K + + AK +R VR + D
Sbjct: 533 NNSNLIGINNGNSESSSTDEESCKKPREE------ENIKAKISRAYVRTEVSDTGLIVKD 694
Query: 180 GCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLP- 238
G WRKYGQK+ + NPCPRAY++C+ APSCPV+K+VQR V+D S+L+ TYEG HNH P
Sbjct: 695 GYHWRKYGQKVTRDNPCPRAYFKCSFAPSCPVKKKVQRSVDDQSMLVATYEGEHNHPQPP 874
Query: 239 --------------LSATAMASTTSAAASMLLSGSSTSNSGSMPS 269
S AS TS AA +++ ST++ S S
Sbjct: 875 QIESTSGSGRSVNHSSVPCSASLTSPAAPKVVTLDSTTSKNSKDS 1009
>TC80880 similar to SP|Q9ZQ70|WRK3_ARATH Probable WRKY transcription factor
3 (WRKY DNA-binding protein 3). [Mouse-ear cress],
partial (24%)
Length = 907
Score = 105 bits (261), Expect = 6e-23
Identities = 63/171 (36%), Positives = 88/171 (50%), Gaps = 2/171 (1%)
Frame = +2
Query: 101 KFETSKSGSTTEGLPNIPSPVNSSEVVPIKNDEVVETW--PPSKTLNKTMRDAEDEVAQQ 158
K E S +T E L S + SE V + EV E P SK N + + +
Sbjct: 11 KMEPESSQATVEHL----SGTSDSEDVGDRETEVHEKRIEPDSKRRNTEVTVSNPTTSSH 178
Query: 159 TPAKKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRC 218
+ ++ V+ + ++DG +WRKYGQK+ KGNP PR+YY+CT P C VRK V+R
Sbjct: 179 RTVTEPKIIVQTTSEVDLLDDGYRWRKYGQKVVKGNPYPRSYYKCT-TPGCNVRKHVERA 355
Query: 219 VEDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPS 269
D +ITTYEG HNH +P + T + + AS L S ++ S S S
Sbjct: 356 STDPKAVITTYEGKHNHDVPAAKTNSHTIANNNASQLKSQNTISEKTSFGS 508
>BG645618 similar to GP|18158619|gb WRKY-like drought-induced protein {Retama
raetam}, partial (37%)
Length = 747
Score = 103 bits (258), Expect = 1e-22
Identities = 68/196 (34%), Positives = 97/196 (48%)
Frame = -1
Query: 58 SLGRVPSNNIPKNDQEKVNKVSKLALNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNI 117
S G V S ++D N+ S LA+ +D+ N E S+ N
Sbjct: 681 SAGSVMSTQGERSD----NRASSLAVRDDKASNSPEQSVVAT----------------ND 562
Query: 118 PSPVNSSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTM 177
SP + V ND V + P SK + +A D + P ++ RV V+ + +
Sbjct: 561 LSPEGAGFVSTRTNDGVDDDDPFSKQRKMELGNA-DIIPVVKPIREPRVVVQTMSEIDIL 385
Query: 178 NDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSL 237
+DG +WRKYGQK+ +GNP PR+YY+CT A CPVRK V+R D +ITTYEG HNH +
Sbjct: 384 DDGYRWRKYGQKVVRGNPNPRSYYKCTNA-GCPVRKHVERASHDPKAVITTYEGKHNHDV 208
Query: 238 PLSATAMASTTSAAAS 253
P + ++ A S
Sbjct: 207 PAARSSSHDMAGHATS 160
>TC88624 similar to PIR|S51529|S51529 SPF1 protein - sweet potato, partial
(50%)
Length = 1496
Score = 101 bits (251), Expect = 8e-22
Identities = 70/220 (31%), Positives = 103/220 (46%), Gaps = 4/220 (1%)
Frame = +2
Query: 23 RALEMQFNNMVKQETKKNNDNNHEEMNAESDLVSLSLGRVPSNNIPKNDQEKVNKVSKLA 82
R++E Q +V + T + N+ S S +L VP N I + +
Sbjct: 881 RSIEGQVTEIVYKGTHNHPKPQCTRRNSSSS--SNALVVVPVNPINEIHDQSYASHGNGQ 1054
Query: 83 LNNDEEFNKEELSLGLECKFETSKSGSTTEGLPNIPSPVNSSEVVPIKNDEVVETWPPSK 142
+++ +S+G + FE S + G DE E P +K
Sbjct: 1055 MDSAATPENSSISIGGDDDFEQSSHQRSRSGGAG---------------DEFDEEEPEAK 1189
Query: 143 TLNKTMRDAEDEVAQQTPAKKA----RVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPR 198
+ + E+E PA + RV V+ D ++DG +WRKYGQK+ KGNP PR
Sbjct: 1190 ---RWKNEGENEGISAQPASRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPR 1360
Query: 199 AYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLP 238
+YY+CT P+CPVRK V+R D+ +ITTYEG HNH +P
Sbjct: 1361 SYYKCT-HPNCPVRKHVERASHDLRAVITTYEGKHNHDVP 1477
Score = 73.2 bits (178), Expect = 2e-13
Identities = 41/108 (37%), Positives = 61/108 (55%), Gaps = 10/108 (9%)
Frame = +2
Query: 178 NDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSL 237
+DG WRKYGQK KG+ PR+YY+CT P+CP +K+V+R +E + Y+GTHNH
Sbjct: 767 DDGYNWRKYGQKQVKGSENPRSYYKCTY-PNCPTKKKVERSIEG-QVTEIVYKGTHNHPK 940
Query: 238 PLSATAMASTTSAAASMLL----------SGSSTSNSGSMPSAQTNNN 275
P T S++S+ A +++ ++ +G M SA T N
Sbjct: 941 P-QCTRRNSSSSSNALVVVPVNPINEIHDQSYASHGNGQMDSAATPEN 1081
>TC78376 similar to SP|Q9SK33|WR60_ARATH Probable WRKY transcription factor
60 (WRKY DNA-binding protein 60). [Mouse-ear cress],
partial (38%)
Length = 1121
Score = 99.8 bits (247), Expect = 2e-21
Identities = 45/92 (48%), Positives = 63/92 (67%), Gaps = 5/92 (5%)
Frame = +1
Query: 177 MNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHS 236
+ DG QWRKYGQK+ + NP PRAY++C+ AP CPV+K+VQR VED ++L+TTYEG HNH+
Sbjct: 589 VKDGYQWRKYGQKVTRDNPSPRAYFKCSFAPGCPVKKKVQRSVEDQNVLVTTYEGEHNHA 768
Query: 237 -----LPLSATAMASTTSAAASMLLSGSSTSN 263
+ L+++ + TT + S SS N
Sbjct: 769 HHQPEMSLTSSNQSETTPTYNLVPASSSSPIN 864
>TC79565 similar to SP|Q9ZQ70|WRK3_ARATH Probable WRKY transcription factor 3
(WRKY DNA-binding protein 3). [Mouse-ear cress], partial
(35%)
Length = 1602
Score = 96.3 bits (238), Expect = 3e-20
Identities = 57/170 (33%), Positives = 79/170 (45%)
Frame = +2
Query: 105 SKSGSTTEGLPNIPSPVNSSEVVPIKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKA 164
SK + + + S + SE EV + P KT + K
Sbjct: 860 SKLTNDNSSVQQVLSGTSDSEEEGDHETEV--DYEPGLKRRKTEAKLLNPALSHRTVSKP 1033
Query: 165 RVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSI 224
++ V+ D + DG +WRKYGQK+ KGNP PR+YY+CT P C VRK V+R D
Sbjct: 1034 KIIVQTTSDVDLLEDGYRWRKYGQKVVKGNPYPRSYYKCT-TPGCNVRKHVERVSTDPKA 1210
Query: 225 LITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNN 274
++TTYEG HNH +P A T S + S S + ++P Q N
Sbjct: 1211 VLTTYEGKHNHDVP-----AAKTNSHNLASNNSASQLKSQNAIPEMQNFN 1345
Score = 69.7 bits (169), Expect = 3e-12
Identities = 51/165 (30%), Positives = 75/165 (44%), Gaps = 2/165 (1%)
Frame = +2
Query: 178 NDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSL 237
+DG WRKYGQK KG PR+YY+CT PSC V K+V+R D + Y+G H H
Sbjct: 674 DDGYNWRKYGQKQVKGCEFPRSYYKCT-HPSCLVTKKVERDPVDGHVTAIIYKGEHIHQR 850
Query: 238 PLSATAMASTTSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNFYLPD-GTKSNQLYLSN 296
P + + +++ +LSG+S S + + Y P + + L N
Sbjct: 851 P--RPSKLTNDNSSVQQVLSGTSDSEEEGDHETEVD--------YEPGLKRRKTEAKLLN 1000
Query: 297 PALSSQH-SHPTITLDLTSNPSNSSTSSPFVRFNSSYNNNNQLPR 340
PALS + S P I + TS+ + ++ N PR
Sbjct: 1001PALSHRTVSKPKIIVQTTSDVDLLEDGYRWRKYGQKVVKGNPYPR 1135
>TC79316 similar to GP|11493822|gb|AAG35658.1 transcription factor WRKY4
{Petroselinum crispum}, partial (49%)
Length = 1282
Score = 95.1 bits (235), Expect = 6e-20
Identities = 52/166 (31%), Positives = 85/166 (50%), Gaps = 11/166 (6%)
Frame = +3
Query: 145 NKTMRDAEDEVAQQTPAKKARVCVRARCD-----------TPTMNDGCQWRKYGQKIAKG 193
N ++++ + + P + C++A+ + + DG QWRKYGQK+ +
Sbjct: 468 NSESSSSDEDQSCKKPRQIVETCIKAKVSRVYFKTESSDSSLIVKDGYQWRKYGQKVTRD 647
Query: 194 NPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLSATAMASTTSAAAS 253
NP PRAY++C+ APSC V+K+VQR V+D S+++ TYEG HNH P + T + +
Sbjct: 648 NPSPRAYFKCSFAPSCTVKKKVQRSVDDPSVVVATYEGEHNHPHP---SQTEPTNNRCRN 818
Query: 254 MLLSGSSTSNSGSMPSAQTNNNLHGLNFYLPDGTKSNQLYLSNPAL 299
+ S + S S PS T + P S + ++NP +
Sbjct: 819 QI---SPVAPSSSAPSTVTLD----WTKKAPSSAPSKNMMINNPKM 935
>TC81471 similar to PIR|A84913|A84913 probable WRKY-type DNA binding protein
[imported] - Arabidopsis thaliana, partial (43%)
Length = 1274
Score = 95.1 bits (235), Expect = 6e-20
Identities = 67/208 (32%), Positives = 101/208 (48%), Gaps = 3/208 (1%)
Frame = +2
Query: 121 VNSSEVVPIKNDEVVETWPPSKT-LNKTMRDAEDEVAQQTPAKKARVCVRARCDTPTMND 179
V S ++ + D+ E K NK ++ + + +Q ++ R + + + D
Sbjct: 392 VKQSMIIKLLVDQAEEDEEEEKQKTNKQLKTKKTNLKRQ---REPRFAFMTKSEVDHLED 562
Query: 180 GCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLP- 238
G +WRKYGQK K +P PR+YYRCT A SC V+K+V+R D SI++TTYEG H H P
Sbjct: 563 GYRWRKYGQKAVKNSPFPRSYYRCTTA-SCNVKKRVERSYTDPSIVVTTYEGQHTHPSPT 739
Query: 239 LSATAMAST-TSAAASMLLSGSSTSNSGSMPSAQTNNNLHGLNFYLPDGTKSNQLYLSNP 297
+S +A A A ++ G ST+N GS+ + H + +Q L N
Sbjct: 740 MSRSAFAGVQIPQPAGVVSGGFSTTNFGSVLQGNYLSQYHQQPY-------QHQQLLVNT 898
Query: 298 ALSSQHSHPTITLDLTSNPSNSSTSSPF 325
S H + N S+S +SPF
Sbjct: 899 LSSLSHPY---------NNSSSFKNSPF 955
>TC88516 similar to GP|14530681|dbj|BAB61053. WRKY DNA-binding protein
{Nicotiana tabacum}, partial (17%)
Length = 1085
Score = 93.6 bits (231), Expect = 2e-19
Identities = 41/77 (53%), Positives = 54/77 (69%)
Frame = +2
Query: 162 KKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVED 221
K+ RV V+ + ++DG +WRKYGQK+ KGNP R+YY+CT AP C VRK V+R D
Sbjct: 236 KEPRVVVQTTSEIDILDDGFRWRKYGQKVVKGNPNARSYYKCT-APGCNVRKHVERAAHD 412
Query: 222 MSILITTYEGTHNHSLP 238
+ +ITTYEG HNH +P
Sbjct: 413 IKAVITTYEGKHNHDVP 463
>TC82987 similar to GP|15991740|gb|AAL13047.1 WRKY transcription factor 71
{Arabidopsis thaliana}, partial (33%)
Length = 975
Score = 93.2 bits (230), Expect = 2e-19
Identities = 58/165 (35%), Positives = 84/165 (50%), Gaps = 11/165 (6%)
Frame = +1
Query: 109 STTEGLPNIPSPVNSSEVVPIKNDEVVE---TWPPSKTLNKTMRDAEDEVAQ-------- 157
S+ G+ P SS N+ V+E T K D +DE ++
Sbjct: 451 SSLVGISETPKSSESSS----SNEAVIEEDSTNKKDKQPKLGCEDGDDEKSKKENKAKKK 618
Query: 158 QTPAKKARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQR 217
+ K+ R + + + DG +WRKYGQK K +P PR+YYRCT + C V+K+V+R
Sbjct: 619 EKKVKEPRFAFLTKSEIDHLEDGYRWRKYGQKAVKNSPYPRSYYRCT-SQKCIVKKRVER 795
Query: 218 CVEDMSILITTYEGTHNHSLPLSATAMASTTSAAASMLLSGSSTS 262
+D SI++TTYEG HNH P + AS+ ++S L SSTS
Sbjct: 796 SYQDPSIVMTTYEGQHNHHCPATLRGNASSMLLSSSPSLFASSTS 930
>TC80708 weakly similar to GP|14530685|dbj|BAB61055. WRKY DNA-binding
protein {Nicotiana tabacum}, partial (20%)
Length = 864
Score = 88.6 bits (218), Expect = 6e-18
Identities = 39/85 (45%), Positives = 59/85 (68%)
Frame = +2
Query: 163 KARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDM 222
++RV VR ++ +NDG +WRKYGQK+ KGN PR YYRC+ +P CPV+K V++ ++
Sbjct: 146 ESRVIVRTTSESGIVNDGYRWRKYGQKMVKGNTNPRNYYRCS-SPGCPVKKHVEKSSQNT 322
Query: 223 SILITTYEGTHNHSLPLSATAMAST 247
+ +ITTYEG H+H+ P + +T
Sbjct: 323 TTVITTYEGQHDHAPPTGRGVLDNT 397
>TC87361 similar to GP|15991736|gb|AAL13045.1 WRKY transcription factor 69
{Arabidopsis thaliana}, partial (40%)
Length = 1452
Score = 88.6 bits (218), Expect = 6e-18
Identities = 48/134 (35%), Positives = 73/134 (53%), Gaps = 8/134 (5%)
Frame = +2
Query: 129 IKNDEVVETWPPSKTLNKTMRDAEDEVAQQTPAKKARVCV--------RARCDTPTMNDG 180
+K + ++E P S + + + E ++ KK V + ++R +T +D
Sbjct: 446 LKLEPILEDGPASPSSCEDTKIEESSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDS 625
Query: 181 CQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLS 240
WRKYGQK KG+P PR YYRC+ + CP RKQV+R D + L+ TY HNHSLPL
Sbjct: 626 WAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLP 805
Query: 241 ATAMASTTSAAASM 254
+ +ST + A++
Sbjct: 806 KSHHSSTNAVTATV 847
>TC80165 similar to PIR|A84913|A84913 probable WRKY-type DNA binding protein
[imported] - Arabidopsis thaliana, partial (32%)
Length = 1013
Score = 85.9 bits (211), Expect = 4e-17
Identities = 48/139 (34%), Positives = 72/139 (51%), Gaps = 20/139 (14%)
Frame = +1
Query: 115 PNIPSPVNSSEVVPIKNDEVVETWPPSKTL------------NKTMRDAEDEVAQQTPAK 162
P + S N E + N + P S ++ NKT+ +++ +Q AK
Sbjct: 385 PPLESDGNGKEYSEVLNSQQQPATPNSSSISSASSEAINDEHNKTVDQTNNQLNKQLKAK 564
Query: 163 K--------ARVCVRARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPSCPVRKQ 214
K AR+ + + + DG +WRKYGQK K +P PR+YYRCT + SC V+K
Sbjct: 565 KTNQKKPREARIAFMTKSEVDHLEDGYRWRKYGQKAVKNSPFPRSYYRCT-SVSCNVKKH 741
Query: 215 VQRCVEDMSILITTYEGTH 233
V+R + D +I++TTYEG H
Sbjct: 742 VERSLSDPTIVVTTYEGKH 798
>TC88395 similar to SP|Q9LP56|WR65_ARATH Probable WRKY transcription factor
65 (WRKY DNA-binding protein 65). [Mouse-ear cress],
partial (43%)
Length = 1086
Score = 84.7 bits (208), Expect = 8e-17
Identities = 45/103 (43%), Positives = 60/103 (57%), Gaps = 1/103 (0%)
Frame = +2
Query: 141 SKTLNKTMRDAEDEVAQQTPAKKARVC-VRARCDTPTMNDGCQWRKYGQKIAKGNPCPRA 199
S + +K R A + Q P K+ ++ +TP +D WRKYGQK KG+P PRA
Sbjct: 140 SSSSSKKSRRAIQKRVVQIPIKEPHGSRLKGESNTPP-SDSWAWRKYGQKPIKGSPYPRA 316
Query: 200 YYRCTVAPSCPVRKQVQRCVEDMSILITTYEGTHNHSLPLSAT 242
YYRC+ CP RKQV+R D ++LI TY HNH+ P+S T
Sbjct: 317 YYRCSSCKGCPARKQVERSRVDPTMLIITYSSDHNHAWPVSKT 445
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.306 0.121 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,173,975
Number of Sequences: 36976
Number of extensions: 244742
Number of successful extensions: 2073
Number of sequences better than 10.0: 161
Number of HSP's better than 10.0 without gapping: 1929
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2016
length of query: 536
length of database: 9,014,727
effective HSP length: 101
effective length of query: 435
effective length of database: 5,280,151
effective search space: 2296865685
effective search space used: 2296865685
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146776.15


