

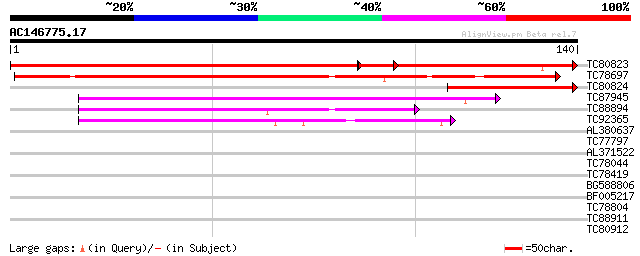
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146775.17 + phase: 0 /pseudo
(140 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80823 similar to GP|13359433|dbj|BAB33412. putative senescence... 176 1e-60
TC78697 similar to GP|13359433|dbj|BAB33412. putative senescence... 162 4e-41
TC80824 69 6e-13
TC87945 weakly similar to GP|17473713|gb|AAL38309.1 unknown prot... 59 8e-10
TC88894 weakly similar to GP|17473713|gb|AAL38309.1 unknown prot... 50 3e-07
TC92365 42 1e-04
AL380637 similar to GP|12698095|dbj hypothetical protein {Macaca... 28 1.2
TC77797 similar to GP|6714354|gb|AAF26045.1| ARG1 protein (Alter... 27 2.1
AL371522 weakly similar to GP|13940621|gb| Putative Mic1 homolog... 27 3.5
TC78044 homologue to SP|P25890|CATA_PEA Catalase (EC 1.11.1.6). ... 26 4.6
TC78419 homologue to GP|6552504|gb|AAF16414.1| mannosyl-oligosac... 26 6.0
BG588806 weakly similar to GP|19920188|g Putative Glucan 1 3-bet... 25 7.9
BF005217 similar to GP|18650643|gb At1g21480/F24J8_23 {Arabidops... 25 7.9
TC78804 25 7.9
TC88911 weakly similar to GP|13926280|gb|AAK49610.1 AT5g14920/F2... 25 7.9
TC80912 similar to SP|O23547|EXR1_ARATH Expansin-related protein... 25 7.9
>TC80823 similar to GP|13359433|dbj|BAB33412. putative senescence-associated
protein {Pisum sativum}, partial (41%)
Length = 794
Score = 176 bits (445), Expect(3) = 1e-60
Identities = 87/87 (100%), Positives = 87/87 (100%)
Frame = +1
Query: 1 MLLSKQKKNQNANNAKRRLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLP 60
MLLSKQKKNQNANNAKRRLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLP
Sbjct: 352 MLLSKQKKNQNANNAKRRLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLP 531
Query: 61 VLGNDHSAFFLYCPHLGSDAALSPWDK 87
VLGNDHSAFFLYCPHLGSDAALSPWDK
Sbjct: 532 VLGNDHSAFFLYCPHLGSDAALSPWDK 612
Score = 68.9 bits (167), Expect(3) = 1e-60
Identities = 37/46 (80%), Positives = 39/46 (84%), Gaps = 1/46 (2%)
Frame = +3
Query: 96 FVLCKKPQAATNEAAAEDGSGTSSLPRRGSGSWKS-WFNLNLKVSS 140
FVLCKKPQAATNEAAAEDGSGTSSL RRG +S WFN +LKVSS
Sbjct: 639 FVLCKKPQAATNEAAAEDGSGTSSLSRRGKW*LESLWFNPHLKVSS 776
Score = 25.4 bits (54), Expect(3) = 1e-60
Identities = 10/10 (100%), Positives = 10/10 (100%)
Frame = +2
Query: 87 KIGSHGARNF 96
KIGSHGARNF
Sbjct: 611 KIGSHGARNF 640
>TC78697 similar to GP|13359433|dbj|BAB33412. putative senescence-associated
protein {Pisum sativum}, partial (41%)
Length = 1058
Score = 162 bits (410), Expect = 4e-41
Identities = 91/136 (66%), Positives = 108/136 (78%), Gaps = 1/136 (0%)
Frame = +1
Query: 2 LLSKQKKNQNANNAKRRLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPV 61
+L KQKKNQNA +A R+LLI+INVLGSAGPIRFVV+E +LV VIDT LKSYAREGRLP+
Sbjct: 346 MLIKQKKNQNAKDA-RKLLININVLGSAGPIRFVVSEGDLVATVIDTALKSYAREGRLPI 522
Query: 62 LGNDHSAFFLYCPHLGSDAALSPWDKIGSH-GARNFVLCKKPQAATNEAAAEDGSGTSSL 120
LG D ++F LYC H+GSD ALSP D IGSH GARNF+LCKKP+ TN A DG+ +L
Sbjct: 523 LGKDVTSFALYCQHVGSD-ALSPLDTIGSHGGARNFMLCKKPE-TTNRMANADGA--VAL 690
Query: 121 PRRGSGSWKSWFNLNL 136
RR +GS K+WFN +L
Sbjct: 691 ARRRNGSLKNWFNKSL 738
>TC80824
Length = 342
Score = 68.9 bits (167), Expect = 6e-13
Identities = 32/32 (100%), Positives = 32/32 (100%)
Frame = +1
Query: 109 AAAEDGSGTSSLPRRGSGSWKSWFNLNLKVSS 140
AAAEDGSGTSSLPRRGSGSWKSWFNLNLKVSS
Sbjct: 1 AAAEDGSGTSSLPRRGSGSWKSWFNLNLKVSS 96
>TC87945 weakly similar to GP|17473713|gb|AAL38309.1 unknown protein
{Arabidopsis thaliana}, partial (48%)
Length = 1087
Score = 58.5 bits (140), Expect = 8e-10
Identities = 35/107 (32%), Positives = 57/107 (52%), Gaps = 3/107 (2%)
Frame = +2
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLGNDHSAFFLYCPHLG 77
+LL+ + VLGS GPI+ ++ E V +++T ++ Y E R P+L + + F
Sbjct: 422 KLLLKVTVLGSLGPIQMLMKPESTVGDLVETAVRQYINEARRPILPSRFGSDFELHFSQF 601
Query: 78 SDAALSPWDKIGSHGARNFVLCKKPQAATNEAAA---EDGSGTSSLP 121
S +L+ +K+ G+RNF LC K A E + DGS T++ P
Sbjct: 602 SLESLNREEKLVKLGSRNFFLCPKNTAHAGEGISGNRNDGSVTTTFP 742
>TC88894 weakly similar to GP|17473713|gb|AAL38309.1 unknown protein
{Arabidopsis thaliana}, partial (48%)
Length = 736
Score = 50.1 bits (118), Expect = 3e-07
Identities = 28/85 (32%), Positives = 48/85 (55%), Gaps = 1/85 (1%)
Frame = +1
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVL-GNDHSAFFLYCPHL 76
+LLI + ++GS GP++ ++ E V +I ++ Y +EGR P+L D S F L+
Sbjct: 412 KLLIKVMMMGSLGPVQVLMTMESTVGDLIAEAVRLYVKEGRRPILPSKDASCFDLHYSQF 591
Query: 77 GSDAALSPWDKIGSHGARNFVLCKK 101
+ +L +K+ G+RNF +C K
Sbjct: 592 SLE-SLDRNEKLREIGSRNFFMCPK 663
>TC92365
Length = 583
Score = 41.6 bits (96), Expect = 1e-04
Identities = 28/96 (29%), Positives = 56/96 (58%), Gaps = 3/96 (3%)
Frame = +2
Query: 18 RLLISINVLGSAGPIRFVVNEEELVEAVIDTTLKSYAREGRLPVLGN-DHSAFFL-YCPH 75
+LL+++++ + G I+ ++ E+ V ++ L +Y +E R P+L + D + + L Y P
Sbjct: 125 KLLLNVSIENTLGAIQVLMLPEDTVGQLVKVALMTYDKEKRKPLLKDTDPNRYQLHYSPF 304
Query: 76 LGSDAALSPWDKIGSHGARNFVLCKKPQAA-TNEAA 110
+S +K+ + G++NF LC KP + T++AA
Sbjct: 305 TLESLKVS--EKLKNLGSKNFFLCSKPSSI*TSKAA 406
>AL380637 similar to GP|12698095|dbj hypothetical protein {Macaca
fascicularis}, partial (3%)
Length = 448
Score = 28.1 bits (61), Expect = 1.2
Identities = 8/34 (23%), Positives = 23/34 (67%)
Frame = -1
Query: 96 FVLCKKPQAATNEAAAEDGSGTSSLPRRGSGSWK 129
F C +++++E+++++ S ++S+P G+W+
Sbjct: 247 FSFCSSDESSSSESSSDESSSSNSMPG*QKGTWR 146
>TC77797 similar to GP|6714354|gb|AAF26045.1| ARG1 protein (Altered Response
to Gravity); 32591-35072 {Arabidopsis thaliana}, partial
(93%)
Length = 2063
Score = 27.3 bits (59), Expect = 2.1
Identities = 16/32 (50%), Positives = 19/32 (59%)
Frame = +2
Query: 108 EAAAEDGSGTSSLPRRGSGSWKSWFNLNLKVS 139
E+ EDG G+ S + G K WFNLNLK S
Sbjct: 1508 ESPGEDG-GSDSKDKSGK---KKWFNLNLKGS 1591
>AL371522 weakly similar to GP|13940621|gb| Putative Mic1 homolog {Oryza
sativa}, partial (5%)
Length = 462
Score = 26.6 bits (57), Expect = 3.5
Identities = 12/34 (35%), Positives = 18/34 (52%)
Frame = +1
Query: 46 IDTTLKSYAREGRLPVLGNDHSAFFLYCPHLGSD 79
I + S R+ RL +L N+H F + C H G +
Sbjct: 28 IHSNKSSQNRDLRLILLHNEHHRFLMLCEHEGKE 129
>TC78044 homologue to SP|P25890|CATA_PEA Catalase (EC 1.11.1.6). [Garden
pea] {Pisum sativum}, partial (56%)
Length = 961
Score = 26.2 bits (56), Expect = 4.6
Identities = 15/46 (32%), Positives = 25/46 (53%), Gaps = 5/46 (10%)
Frame = +3
Query: 32 IRF--VVNEEELVEAVIDT---TLKSYAREGRLPVLGNDHSAFFLY 72
+RF V++E E + D +K Y REG ++GN+ FF++
Sbjct: 426 VRFSTVIHERGSPETLRDPRGFAVKFYTREGNFDLVGNNFPVFFVH 563
>TC78419 homologue to GP|6552504|gb|AAF16414.1| mannosyl-oligosaccharide 1
2-alpha-mannosidase {Glycine max}, partial (79%)
Length = 1832
Score = 25.8 bits (55), Expect = 6.0
Identities = 13/36 (36%), Positives = 19/36 (52%)
Frame = +2
Query: 39 EELVEAVIDTTLKSYAREGRLPVLGNDHSAFFLYCP 74
++ VE VI K++ +G LP+ N HS Y P
Sbjct: 488 QQKVENVITVLNKTFPDDGLLPIYINPHSGTTGYSP 595
>BG588806 weakly similar to GP|19920188|g Putative Glucan 1
3-beta-glucosidase precursor {Oryza sativa (japonica
cultivar-group)}, partial (9%)
Length = 791
Score = 25.4 bits (54), Expect = 7.9
Identities = 12/25 (48%), Positives = 15/25 (60%)
Frame = +3
Query: 110 AAEDGSGTSSLPRRGSGSWKSWFNL 134
+AEDG GT+ + RGS S F L
Sbjct: 153 SAEDGGGTTIVANRGSASGWETFRL 227
>BF005217 similar to GP|18650643|gb At1g21480/F24J8_23 {Arabidopsis
thaliana}, partial (40%)
Length = 624
Score = 25.4 bits (54), Expect = 7.9
Identities = 8/28 (28%), Positives = 17/28 (60%)
Frame = -3
Query: 59 LPVLGNDHSAFFLYCPHLGSDAALSPWD 86
+P+ G ++ A ++ CP L ++ S W+
Sbjct: 466 IPLRGKNNGAIYICCPRLEQVSSSSTWE 383
>TC78804
Length = 1318
Score = 25.4 bits (54), Expect = 7.9
Identities = 11/42 (26%), Positives = 23/42 (54%)
Frame = +3
Query: 37 NEEELVEAVIDTTLKSYAREGRLPVLGNDHSAFFLYCPHLGS 78
N +L + I+ + +++GRL ++ FF++ PH+ S
Sbjct: 591 NTSQLADVFINLSENFESKQGRLHERSSNLVEFFIHLPHIRS 716
>TC88911 weakly similar to GP|13926280|gb|AAK49610.1 AT5g14920/F2G14_40
{Arabidopsis thaliana}, partial (11%)
Length = 1320
Score = 25.4 bits (54), Expect = 7.9
Identities = 17/49 (34%), Positives = 21/49 (42%)
Frame = -3
Query: 63 GNDHSAFFLYCPHLGSDAALSPWDKIGSHGARNFVLCKKPQAATNEAAA 111
G+D AFF C G+ +LS GA F LC A + A A
Sbjct: 730 GDDFGAFFFLCVGAGAGVSLS------GAGAGAFFLCVGAGAGASLAGA 602
>TC80912 similar to SP|O23547|EXR1_ARATH Expansin-related protein 1
precursor (At-EXPR1) (Ath-ExpBeta-3.1). [Mouse-ear
cress], partial (35%)
Length = 764
Score = 25.4 bits (54), Expect = 7.9
Identities = 11/30 (36%), Positives = 16/30 (52%)
Frame = -2
Query: 59 LPVLGNDHSAFFLYCPHLGSDAALSPWDKI 88
LP L + + ++CP + S A SPW I
Sbjct: 295 LP*LRHTSNIAIIHCPSIFSKATSSPWSPI 206
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.314 0.131 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,493,547
Number of Sequences: 36976
Number of extensions: 56035
Number of successful extensions: 245
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 238
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 239
length of query: 140
length of database: 9,014,727
effective HSP length: 86
effective length of query: 54
effective length of database: 5,834,791
effective search space: 315078714
effective search space used: 315078714
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 53 (25.0 bits)
Medicago: description of AC146775.17


