

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146775.16 - phase: 0
(172 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
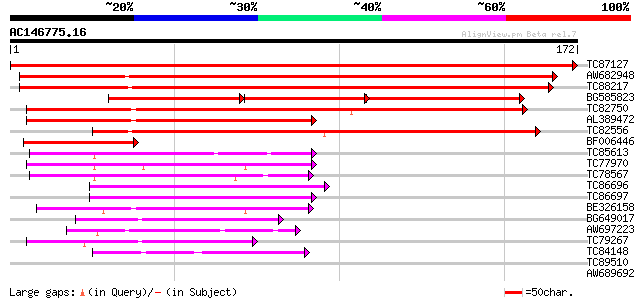
Score E
Sequences producing significant alignments: (bits) Value
TC87127 similar to GP|12324745|gb|AAG52327.1 unknown protein; 38... 352 4e-98
AW682948 weakly similar to PIR|E96525|E96 protein T1N15.21 [impo... 223 2e-59
TC88217 weakly similar to GP|18377801|gb|AAL67050.1 unknown prot... 212 7e-56
BG585823 similar to GP|12324745|gb| unknown protein; 3866-2463 {... 99 2e-40
TC82750 similar to GP|21592759|gb|AAM64708.1 unknown {Arabidopsi... 111 2e-25
AL389472 similar to GP|6648206|gb|A putative GTPase activating p... 100 3e-22
TC82556 similar to GP|21536965|gb|AAM61306.1 putative zinc finge... 97 3e-21
BF006446 similar to PIR|E96525|E965 protein T1N15.21 [imported] ... 71 2e-13
TC85613 weakly similar to EGAD|2627|2550 protein kinase C beta ... 57 4e-09
TC77970 similar to GP|20197686|gb|AAM15203.1 expressed protein {... 51 2e-07
TC78567 similar to PIR|T04143|T04143 CLB1 protein - tomato, part... 47 5e-06
TC86696 similar to GP|3860331|emb|CAA10133.1 hypothetical protei... 47 5e-06
TC86697 homologue to GP|3860331|emb|CAA10133.1 hypothetical prot... 45 1e-05
BE326158 similar to GP|10177757|d phosphoribosylanthranilate tra... 43 5e-05
BG649017 similar to GP|9757890|db phosphoribosylanthranilate tra... 42 1e-04
AW697223 similar to GP|9757890|dbj phosphoribosylanthranilate tr... 41 3e-04
TC79267 similar to PIR|T00782|T00782 probable anthranilate phosp... 40 6e-04
TC84148 similar to GP|11994100|dbj|BAB01103. gene_id:MYF24.9~unk... 39 8e-04
TC89510 weakly similar to GP|22831331|dbj|BAC16176. contains EST... 37 0.003
AW689692 weakly similar to GP|22795064|gb phospholipase D delta ... 33 0.057
>TC87127 similar to GP|12324745|gb|AAG52327.1 unknown protein; 3866-2463
{Arabidopsis thaliana}, partial (76%)
Length = 747
Score = 352 bits (903), Expect = 4e-98
Identities = 172/172 (100%), Positives = 172/172 (100%)
Frame = +3
Query: 1 MDNNVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTL 60
MDNNVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTL
Sbjct: 24 MDNNVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTL 203
Query: 61 SIRDVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNGCAIKRVQANRT 120
SIRDVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNGCAIKRVQANRT
Sbjct: 204 SIRDVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNGCAIKRVQANRT 383
Query: 121 NCLAEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKGLLGAQM 172
NCLAEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKGLLGAQM
Sbjct: 384 NCLAEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKGLLGAQM 539
>AW682948 weakly similar to PIR|E96525|E96 protein T1N15.21 [imported] -
Arabidopsis thaliana, partial (72%)
Length = 576
Score = 223 bits (569), Expect = 2e-59
Identities = 108/163 (66%), Positives = 132/163 (80%)
Frame = +3
Query: 4 NVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIR 63
N+LGL++LRIK+G NL DS +SDPYV+V + EQ LKT VV +NC+PEWNEELTL I+
Sbjct: 12 NILGLIRLRIKKGTNLIPHDSRTSDPYVLVTM-EEQTLKTAVVNDNCHPEWNEELTLYIK 188
Query: 64 DVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNGCAIKRVQANRTNCL 123
DV PI L V DKDTF VDDKMG+A+ID+KPY QCVKM L LP+G +K VQ + TNCL
Sbjct: 189 DVNTPIHLIVCDKDTFTVDDKMGEADIDIKPYLQCVKMGLSDLPDGHVVKTVQPDTTNCL 368
Query: 124 AEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCKG 166
AEESSC+W++GKV+QEM LRLRNVESGE++VEIEW+DV +G
Sbjct: 369 AEESSCVWRDGKVVQEMSLRLRNVESGEVLVEIEWIDVTDSEG 497
>TC88217 weakly similar to GP|18377801|gb|AAL67050.1 unknown protein
{Arabidopsis thaliana}, partial (94%)
Length = 857
Score = 212 bits (539), Expect = 7e-56
Identities = 101/162 (62%), Positives = 127/162 (78%)
Frame = +3
Query: 4 NVLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIR 63
N+LGLLK+ ++RG+NLAIRD SSDPYVV+ + +QKLKTRVVK N NPEWNE+LTLSI
Sbjct: 87 NLLGLLKIHVQRGVNLAIRDVVSSDPYVVIKMA-KQKLKTRVVKKNLNPEWNEDLTLSIS 263
Query: 64 DVRVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNGCAIKRVQANRTNCL 123
D PI L V+DKDTF +DDKMGDAE D+ P+ + VKM+L LPN + RVQ +R NCL
Sbjct: 264 DPHTPIHLYVYDKDTFSLDDKMGDAEFDIGPFFEAVKMRLAGLPNEAIVTRVQPSRQNCL 443
Query: 124 AEESSCIWKNGKVLQEMILRLRNVESGELVVEIEWVDVPGCK 165
AEES +WK+GK+ Q M+LRLRNVE GE+ +++ WVD+PG K
Sbjct: 444 AEESHIVWKDGKIFQNMVLRLRNVECGEVELQLHWVDIPGSK 569
>BG585823 similar to GP|12324745|gb| unknown protein; 3866-2463 {Arabidopsis
thaliana}, partial (51%)
Length = 717
Score = 99.0 bits (245), Expect(2) = 2e-40
Identities = 48/48 (100%), Positives = 48/48 (100%)
Frame = +1
Query: 109 GCAIKRVQANRTNCLAEESSCIWKNGKVLQEMILRLRNVESGELVVEI 156
GCAIKRVQANRTNCLAEESSCIWKNGKVLQEMILRLRNVESGELVVEI
Sbjct: 574 GCAIKRVQANRTNCLAEESSCIWKNGKVLQEMILRLRNVESGELVVEI 717
Score = 83.6 bits (205), Expect(2) = 2e-40
Identities = 38/38 (100%), Positives = 38/38 (100%)
Frame = +3
Query: 72 TVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNG 109
TVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNG
Sbjct: 462 TVFDKDTFFVDDKMGDAEIDLKPYTQCVKMKLDTLPNG 575
Score = 73.6 bits (179), Expect = 4e-14
Identities = 34/41 (82%), Positives = 36/41 (86%)
Frame = +3
Query: 31 VVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDVRVPICL 71
++V QKLKTRVVKNNCNPEWNEELTLSIRDVRVPICL
Sbjct: 48 IIVFFSLMQKLKTRVVKNNCNPEWNEELTLSIRDVRVPICL 170
>TC82750 similar to GP|21592759|gb|AAM64708.1 unknown {Arabidopsis
thaliana}, partial (95%)
Length = 623
Score = 111 bits (277), Expect = 2e-25
Identities = 63/154 (40%), Positives = 94/154 (60%), Gaps = 2/154 (1%)
Frame = +3
Query: 6 LGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDV 65
LG LK+ + +G L IRD +SDPYVV+ +G+ Q KT+V+ + NP WNEEL ++ +
Sbjct: 75 LGQLKVIVVQGKRLVIRDFKTSDPYVVLKLGN-QTAKTKVINSCLNPVWNEELNFTLTEP 251
Query: 66 RVPICLTVFDKDTFFVDDKMGDAEIDLKPYTQCVKMK--LDTLPNGCAIKRVQANRTNCL 123
+ L VFDKD DDKMG+A I+L+P +++ L +++V + NCL
Sbjct: 252 LGVLNLEVFDKDLLKADDKMGNAFINLQPLVSAARLRDILRVSSGEQTLRKVIPDSENCL 431
Query: 124 AEESSCIWKNGKVLQEMILRLRNVESGELVVEIE 157
ESS NG+V+Q + LRLR VESGE+ + ++
Sbjct: 432 VRESSINCVNGEVVQNVWLRLREVESGEIELTLK 533
>AL389472 similar to GP|6648206|gb|A putative GTPase activating protein
{Arabidopsis thaliana}, partial (24%)
Length = 487
Score = 100 bits (249), Expect = 3e-22
Identities = 48/88 (54%), Positives = 68/88 (76%)
Frame = +1
Query: 6 LGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDV 65
+GL+K+ + +G NLAIRD +SDPYV++++GH Q +KTRV+KNN NP WNE L LSI +
Sbjct: 226 VGLIKVNVVKGTNLAIRDIVTSDPYVILSLGH-QSVKTRVIKNNLNPVWNESLMLSIPEN 402
Query: 66 RVPICLTVFDKDTFFVDDKMGDAEIDLK 93
P+ + V+DKD+F DD MG+AEID++
Sbjct: 403 IPPLKIIVYDKDSFKNDDFMGEAEIDIQ 486
>TC82556 similar to GP|21536965|gb|AAM61306.1 putative zinc finger and C2
domain protein {Arabidopsis thaliana}, partial (40%)
Length = 756
Score = 97.1 bits (240), Expect = 3e-21
Identities = 57/137 (41%), Positives = 84/137 (60%), Gaps = 1/137 (0%)
Frame = +3
Query: 26 SSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDVRVPICLTVFDKDTFFVDDKM 85
SSDPYVV+N+G Q ++T V+++N NP WNEE LS+ + + L VFD DTF DD M
Sbjct: 39 SSDPYVVLNLG-TQTVQTSVMRSNLNPVWNEEHMLSVPEHYGQLKLKVFDHDTFSADDIM 215
Query: 86 GDAEIDLKP-YTQCVKMKLDTLPNGCAIKRVQANRTNCLAEESSCIWKNGKVLQEMILRL 144
G+A+IDL+ T + + I + + N L E+S+ +GKV Q M L+L
Sbjct: 216 GEADIDLQSLITSAMAFGDAGMFGDMQIGKWLKSDDNALIEDSAVKIIDGKVKQMMTLKL 395
Query: 145 RNVESGELVVEIEWVDV 161
+NVE GE+ +E+EW+ +
Sbjct: 396 QNVECGEIELELEWISL 446
>BF006446 similar to PIR|E96525|E965 protein T1N15.21 [imported] -
Arabidopsis thaliana, partial (14%)
Length = 575
Score = 71.2 bits (173), Expect = 2e-13
Identities = 35/35 (100%), Positives = 35/35 (100%)
Frame = +2
Query: 5 VLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQ 39
VLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQ
Sbjct: 2 VLGLLKLRIKRGINLAIRDSNSSDPYVVVNIGHEQ 106
>TC85613 weakly similar to EGAD|2627|2550 protein kinase C beta {Homo
sapiens}, partial (3%)
Length = 584
Score = 57.0 bits (136), Expect = 4e-09
Identities = 36/88 (40%), Positives = 49/88 (54%), Gaps = 1/88 (1%)
Frame = +1
Query: 7 GLLKLRIKRGINLAIRDS-NSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDV 65
G LK+ + RG L D+ SDPYV + I + K +T K NPEWNE T ++ D
Sbjct: 85 GTLKVIVVRGKKLKDEDAIGKSDPYVELWIKKDYKQRTTTKKGTVNPEWNETFTFNV-DG 261
Query: 66 RVPICLTVFDKDTFFVDDKMGDAEIDLK 93
+ L V D D DDK+G+A++DLK
Sbjct: 262 DHHLYLKVLDDDV-VSDDKIGEAKVDLK 342
>TC77970 similar to GP|20197686|gb|AAM15203.1 expressed protein {Arabidopsis
thaliana}, partial (97%)
Length = 2062
Score = 51.2 bits (121), Expect = 2e-07
Identities = 32/92 (34%), Positives = 49/92 (52%), Gaps = 4/92 (4%)
Frame = +2
Query: 6 LGLLKLRIKRGINLAIRDS-NSSDPYVVVNIGHEQ--KLKTRVVKNNCNPEWNEELTLSI 62
+G+L +++ + L +D +SDPYV + + ++ KT V N NPEWNEE L +
Sbjct: 947 VGILHVKVLHAMKLKKKDLLGASDPYVKLKLTDDKMPSKKTTVKHKNLNPEWNEEFNLVV 1126
Query: 63 RDVRVPIC-LTVFDKDTFFVDDKMGDAEIDLK 93
+D + L V+D + DKMG I LK
Sbjct: 1127 KDPETQVLQLNVYDWEQVGKHDKMGMNVITLK 1222
>TC78567 similar to PIR|T04143|T04143 CLB1 protein - tomato, partial (53%)
Length = 1159
Score = 46.6 bits (109), Expect = 5e-06
Identities = 31/88 (35%), Positives = 47/88 (53%), Gaps = 2/88 (2%)
Frame = +3
Query: 7 GLLKLRIKRGINLAIRDS-NSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDV 65
G LK+ I + +L + SDPYVV+ I K+KT+V+ NN NP W++ L D
Sbjct: 174 GSLKVTIVKATDLKNMEMIGKSDPYVVLYIRPLFKVKTKVINNNLNPVWDQTFELIAEDK 353
Query: 66 RV-PICLTVFDKDTFFVDDKMGDAEIDL 92
+ L VFD+D D ++G ++ L
Sbjct: 354 ETQSLILEVFDED-IGQDKRLGIVKLPL 434
>TC86696 similar to GP|3860331|emb|CAA10133.1 hypothetical protein {Cicer
arietinum}, partial (90%)
Length = 733
Score = 46.6 bits (109), Expect = 5e-06
Identities = 21/73 (28%), Positives = 36/73 (48%)
Frame = +1
Query: 25 NSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDVRVPICLTVFDKDTFFVDDK 84
+S DPYV++ ++ T NP+WNE ++ D + L + +KD + DD
Sbjct: 151 SSIDPYVILTYRAQEHKSTVQEGAGSNPQWNETFLFTVSDTAYELNLKIMEKDNYSADDN 330
Query: 85 MGDAEIDLKPYTQ 97
+G+ I L+ Q
Sbjct: 331 LGEVIIPLETVIQ 369
>TC86697 homologue to GP|3860331|emb|CAA10133.1 hypothetical protein {Cicer
arietinum}, partial (89%)
Length = 825
Score = 45.1 bits (105), Expect = 1e-05
Identities = 21/69 (30%), Positives = 37/69 (53%)
Frame = +2
Query: 25 NSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDVRVPICLTVFDKDTFFVDDK 84
+S DPYV+++ ++ T NP+WNE ++ D + L + +KD + DD
Sbjct: 203 SSIDPYVILSYSGQEHKSTVQEGAGSNPQWNETFLFTVSDNASELNLKIMEKDNYNNDDN 382
Query: 85 MGDAEIDLK 93
+G+A I L+
Sbjct: 383 IGEAIIPLE 409
>BE326158 similar to GP|10177757|d phosphoribosylanthranilate
transferase-like protein {Arabidopsis thaliana}, partial
(15%)
Length = 618
Score = 43.1 bits (100), Expect = 5e-05
Identities = 29/86 (33%), Positives = 45/86 (51%), Gaps = 2/86 (2%)
Frame = +3
Query: 9 LKLRIKRGINLAIRDSNSS-DPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDVRV 67
L +R+ + NL + S+ DPYV V +G+ K +T+ + NPEWN+ S ++
Sbjct: 177 LYVRVVKAKNLTLNSLTSTCDPYVEVRLGN-YKGRTKHLDKRSNPEWNQVYAFSKDQIQS 353
Query: 68 PIC-LTVFDKDTFFVDDKMGDAEIDL 92
I + V DK+T DD +G DL
Sbjct: 354 SILEVIVKDKETVGRDDYIGRVAFDL 431
>BG649017 similar to GP|9757890|db phosphoribosylanthranilate
transferase-like protein {Arabidopsis thaliana}, partial
(15%)
Length = 554
Score = 42.0 bits (97), Expect = 1e-04
Identities = 20/63 (31%), Positives = 34/63 (53%)
Frame = +3
Query: 21 IRDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDVRVPICLTVFDKDTFF 80
+ +S+D Y V G++ ++TR V +N P+WNE+ T + D + + VFD + F
Sbjct: 96 VNGKSSTDGYCVAKYGNKW-VRTRTVSDNLEPKWNEQYTWKVFDPSTVLTIGVFDSFSVF 272
Query: 81 VDD 83
D
Sbjct: 273 ESD 281
>AW697223 similar to GP|9757890|dbj phosphoribosylanthranilate
transferase-like protein {Arabidopsis thaliana}, partial
(5%)
Length = 778
Score = 40.8 bits (94), Expect = 3e-04
Identities = 27/72 (37%), Positives = 43/72 (59%), Gaps = 1/72 (1%)
Frame = +3
Query: 18 NLAIRDSN-SSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDVRVPICLTVFDK 76
NLA +D + +S PY+V++ H Q+ KTR + + NP WNE L+ ++ + R I V++
Sbjct: 183 NLAPKDGHGTSSPYIVIDF-HGQRRKTRTLVRDLNPVWNETLSFNVGE-RNEIFGDVWEL 356
Query: 77 DTFFVDDKMGDA 88
D + D K G A
Sbjct: 357 DVYH-DMKHGPA 389
>TC79267 similar to PIR|T00782|T00782 probable anthranilate
phosphoribosyltransferase (EC 2.4.2.18) T22J18.21 -
Arabidopsis thaliana, partial (60%)
Length = 1612
Score = 39.7 bits (91), Expect = 6e-04
Identities = 23/73 (31%), Positives = 39/73 (52%), Gaps = 3/73 (4%)
Frame = +3
Query: 6 LGLLKLRIKRGINLAI---RDSNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSI 62
+G L+L I NL +D ++D Y V G++ ++TR + + +P WNE+ T +
Sbjct: 201 IGYLELGILSARNLLPMKGKDGRTTDAYCVAKYGNKW-VRTRTLLDTLSPRWNEQYTWEV 377
Query: 63 RDVRVPICLTVFD 75
D I ++VFD
Sbjct: 378 HDPCTVITVSVFD 416
>TC84148 similar to GP|11994100|dbj|BAB01103. gene_id:MYF24.9~unknown
protein {Arabidopsis thaliana}, partial (16%)
Length = 840
Score = 39.3 bits (90), Expect = 8e-04
Identities = 22/66 (33%), Positives = 36/66 (54%)
Frame = +1
Query: 26 SSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDVRVPICLTVFDKDTFFVDDKM 85
+SDP+V VN G+ +K +T+VV NP W++ TL D P+ L V D + +
Sbjct: 172 TSDPFVRVNYGNLKK-RTKVVHKTINPRWDQ--TLEFLDDGSPLTLHVKDHNALLPTSSI 342
Query: 86 GDAEID 91
G+ ++
Sbjct: 343 GECVVE 360
>TC89510 weakly similar to GP|22831331|dbj|BAC16176. contains ESTs
AU165964(S1723) D40008(S1723)~phosphoribosylanthranilate
transferase-like protein, partial (21%)
Length = 1195
Score = 37.4 bits (85), Expect = 0.003
Identities = 27/86 (31%), Positives = 44/86 (50%), Gaps = 2/86 (2%)
Frame = +3
Query: 9 LKLRIKRGINLAIRD-SNSSDPYVVVNIGHEQKLKTRVVKNNCNPEWNEELTLSIRDVR- 66
L +R+ + +L D + S DPYV+V +G+ + KNN +PEWN + + +
Sbjct: 834 LFIRVVKARDLPRMDLTGSLDPYVIVKVGNFKGTTNHFEKNN-SPEWNLVFAFAKENQQA 1010
Query: 67 VPICLTVFDKDTFFVDDKMGDAEIDL 92
+ + + DKDT DD +G DL
Sbjct: 1011TTLEVVIKDKDTIH-DDFVGTVRFDL 1085
>AW689692 weakly similar to GP|22795064|gb phospholipase D delta isoform 1a
{Gossypium hirsutum}, partial (20%)
Length = 581
Score = 33.1 bits (74), Expect = 0.057
Identities = 18/51 (35%), Positives = 27/51 (52%)
Frame = +1
Query: 43 TRVVKNNCNPEWNEELTLSIRDVRVPICLTVFDKDTFFVDDKMGDAEIDLK 93
TRV+KN+ NPEW E + + + + + V D D F + MG +I K
Sbjct: 94 TRVLKNSLNPEWKERFHIPLAHPVIDLEIRVKDDDVFGA-EVMGMVKIPAK 243
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.138 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,484,272
Number of Sequences: 36976
Number of extensions: 71895
Number of successful extensions: 345
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 335
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 336
length of query: 172
length of database: 9,014,727
effective HSP length: 89
effective length of query: 83
effective length of database: 5,723,863
effective search space: 475080629
effective search space used: 475080629
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC146775.16


