

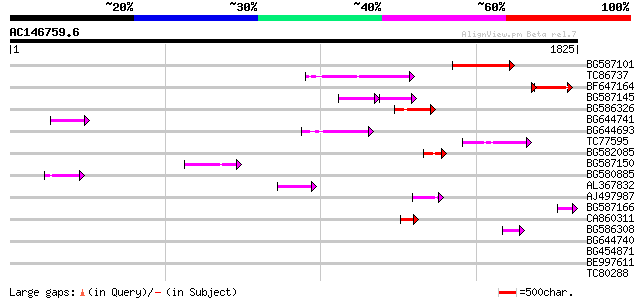
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146759.6 - phase: 0
(1825 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 255 1e-67
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 186 9e-47
BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis t... 151 6e-42
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 91 5e-27
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 92 2e-18
BG644741 90 7e-18
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 86 1e-16
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 69 1e-11
BG582085 69 2e-11
BG587150 similar to GP|13592175|gb ppg3 {Leishmania major}, part... 68 3e-11
BG580885 59 1e-08
AL367832 weakly similar to GP|14091845|gb Putative retroelement ... 54 4e-07
AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis... 53 1e-06
BG587166 similar to GP|8778801|gb| T32E20.9 {Arabidopsis thalian... 51 4e-06
CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product ... 50 6e-06
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 50 8e-06
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 40 0.006
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 38 0.041
BE997611 36 0.12
TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein... 35 0.27
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 255 bits (652), Expect = 1e-67
Identities = 115/198 (58%), Positives = 140/198 (70%)
Frame = +2
Query: 1426 KDANHYLWDDPYLFKVSTDGLIRRCVAGEEIKNIVWHCHSSAYGGHHSGERTAAKVLQSG 1485
KD YLWD+P+L+K D + RCVA EEI I++HCH S Y GH + +T +K+ Q+G
Sbjct: 5 KDVRRYLWDEPFLYKQCADNIYIRCVAEEEIPGILFHCHGSNYAGHFAVSKTVSKIQQAG 184
Query: 1486 FWWPTLFKDCHDFVRRCDNCQRTGSISKRNEMPLTGIIEVEPFDCWGIDFMGPFPPSSSY 1545
FWWPT+FKD H F+ +CD CQR G+IS RNEMP I+EVE FD WGIDFMGPFP S +
Sbjct: 185 FWWPTMFKDAHSFISKCDPCQRQGNIS*RNEMPQNFILEVEVFDVWGIDFMGPFPSSYNN 364
Query: 1546 LHILVCVDYVTKWVEAIPCVANDSKTVVNFLRKNIFTRFGTPRVLISDGGKHFCNNFLET 1605
+ILV VDYV+KWVEAI ND+ VV + IF RFG PRV+ISDGG HF N E
Sbjct: 365 KYILVAVDYVSKWVEAIASPTNDATVVVKMFKSVIFPRFGVPRVVISDGGSHFINKVFEK 544
Query: 1606 VLKKYNIKHKVATPYHPQ 1623
+LKK ++HKVAT YHPQ
Sbjct: 545 LLKKNGVRHKVATAYHPQ 598
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 186 bits (471), Expect = 9e-47
Identities = 120/355 (33%), Positives = 186/355 (51%), Gaps = 4/355 (1%)
Frame = +1
Query: 951 VVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGMTVVVNEKNELIPTRTVTGRRMCID 1010
V+KK + LLD G I S S+ +PV V K GG G R C+D
Sbjct: 508 VLKKTLEDLLDKGFI-KASGSAAGAPVLFVRKPGG------------------GIRFCVD 630
Query: 1011 YRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPEDQEKTAFTCPFG 1070
YR LN T+KD +PLP + + + R+AG ++ LD + ++++ + EDQEKTAF +G
Sbjct: 631 YRALNAITKKDRYPLPLISETLRRVAGARWFTKLDVVAAFHKMRIKDEDQEKTAFRTRYG 810
Query: 1071 VFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVF--GKSFDQCLFHLNAV 1128
+F + PFGL GAPATFQR + + ++ + ++DD ++ G D + V
Sbjct: 811 LFEWIVCPFGLTGAPATFQRYINKTLHEFLDDFVTAYIDDVLIYTTGSKKDH-EAQVRRV 987
Query: 1129 LKRCTETNLILNWEKCHFMVTEGIVLGHKISS-KGIEVDQAKIEVIEKLPPPMNVKGVRS 1187
L+R + L L+ +KC F VT +G +++ KG+ D K+ I PP +VKG RS
Sbjct: 988 LRRLADAGLSLDPKKCEFSVTTVKYVGFILTAGKGVSCDPLKLAAIRDWLPPGSVKGARS 1167
Query: 1188 FLGHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLNAFSLIKNKLVTAPIIIAPNW 1247
FLG +Y+ FI +S+I +PL L K+ F + E AF+ +K P++ +
Sbjct: 1168 FLGFCNYYKDFIPGYSEITEPLTRLTRKDFPFRWGAEQEAAFTKLKRLFAEEPVLRMFDP 1347
Query: 1248 DLHFELMCDASDYAVGAVLGQRKNK-FFHAIYYASKVLNESQVNYSTTEKELLAV 1301
+ + D S +A+G VL Q H + + S+ L+ ++ NY +KELLAV
Sbjct: 1348 EAVTTVETDCSGFALGGVLTQEDGTGAAHPVAFHSQRLSPAEYNYPIHDKELLAV 1512
>BF647164 weakly similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana},
partial (6%)
Length = 469
Score = 151 bits (382), Expect(2) = 6e-42
Identities = 77/125 (61%), Positives = 93/125 (73%)
Frame = +1
Query: 1688 LEHKAYWAIKALNFDQTLAGKKRLLKLNELEEMRLGAYENAVIYKERTKRYHDKGLVRRE 1747
L K IKALN D AG KR+L+LNELEE+RL AYENA IYKERTK++HD+ ++RRE
Sbjct: 28 LSIKPIGPIKALNMDYKAAGGKRVLELNELEELRLDAYENAKIYKERTKKWHDRRIIRRE 207
Query: 1748 FYVGQLVLLFNSRLKLFPGKLKSKWSGPFMIESISPYGAVELSKPGEPGTFKVNAQRIKP 1807
F G+LVLLFNSRLKLFPGKL+S WSGPF ++++ P GAVE+ G F VN QR+K
Sbjct: 208 FREGELVLLFNSRLKLFPGKLRSHWSGPFQVKNVMPSGAVEVWSE-STGPFTVNGQRLKH 384
Query: 1808 YLGGE 1812
Y GE
Sbjct: 385 YTSGE 399
Score = 39.7 bits (91), Expect(2) = 6e-42
Identities = 15/16 (93%), Positives = 16/16 (99%)
Frame = +3
Query: 1680 KACHLPVELEHKAYWA 1695
K+CHLPVELEHKAYWA
Sbjct: 3 KSCHLPVELEHKAYWA 50
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 90.5 bits (223), Expect(2) = 5e-27
Identities = 52/134 (38%), Positives = 73/134 (53%)
Frame = +2
Query: 1057 PEDQEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGK 1116
P+D EKTAF G + Y+ MPFGL A +T+QR + +F+D + +EV++DD V
Sbjct: 8 PDDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVKSL 187
Query: 1117 SFDQCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAKIEVIEKL 1176
L HL K E + LN KC F VT G LG+ ++ +GIEV+ +I I L
Sbjct: 188 RATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAILDL 367
Query: 1177 PPPMNVKGVRSFLG 1190
P P N + V+ G
Sbjct: 368 PSPKNSREVQRLTG 409
Score = 50.8 bits (120), Expect(2) = 5e-27
Identities = 35/118 (29%), Positives = 48/118 (40%)
Frame = +3
Query: 1190 GHAGFYRRFIKDFSKIAKPLCNLLVKETEFDFDVECLNAFSLIKNKLVTAPIIIAPNWDL 1249
G RFI + P LL F +D +C AF +K L T P++ P
Sbjct: 408 GRIAALNRFISRSTDKCLPFYKLLCGNKRFVWDEKCEEAFEQLKQYLTTPPVLSKPEAGD 587
Query: 1250 HFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNESQVNYSTTEKELLAVIFALEK 1307
L S AV +VL + I+Y SK + + + Y T EK AVI + K
Sbjct: 588 TLSLYIAISSTAVSSVLIREDRGEQKPIFYTSKRMTDPETRYPTLEKMAFAVITSARK 761
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 92.0 bits (227), Expect = 2e-18
Identities = 53/132 (40%), Positives = 81/132 (61%)
Frame = +2
Query: 1238 TAPIIIAPNWDLHFELMCDASDYAVGAVLGQRKNKFFHAIYYASKVLNESQVNYSTTEKE 1297
+API++ P + + + DAS +G VL Q + I YAS+ L + + NY T + E
Sbjct: 11 SAPILVLPEL-ITYVVYTDASITGLGCVLTQHEK----VIAYASRQLRKHEGNYPTHDLE 175
Query: 1298 LLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIRDKK 1357
+ AV+FAL+ +RSYL G+KV + TDH +LKY+ T+ + R RW+ + ++DL+I
Sbjct: 176 MAAVVFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYP 355
Query: 1358 GVENVVADHLSR 1369
G N+VAD LSR
Sbjct: 356 GKANLVADALSR 391
>BG644741
Length = 735
Score = 90.1 bits (222), Expect = 7e-18
Identities = 44/128 (34%), Positives = 70/128 (54%)
Frame = -2
Query: 130 CEFCVPPANVNEDQKKLRLFPFTLIGKAKDWLLTLPNGTIQTWEELELKFLEKYFPMSKY 189
C+ CV ++ + LR+FP +L+G+A W LP +I TW +L FL +Y+P+SK
Sbjct: 635 CKSCVGRPYLDLNVLGLRVFPLSLMGEADIWFTELPYNSIFTWNQLRDVFLARYYPVSKK 456
Query: 190 LDKKQEIASFRQGEGESLYDAWERFNLLLKRCPGHEFSEKQYLQFFTEGLTHSNRMFLDA 249
L+ + +F GES+ +W+RF L+ P H + ++F G +N++ LD
Sbjct: 455 LNHNDRVNNFVALPGESVSSSWDRFTSFLRSVPNHRIDDDSLKEYFYRGQDDNNKVVLDT 276
Query: 250 SAGGSLRV 257
AGGS V
Sbjct: 275 IAGGSYGV 252
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 86.3 bits (212), Expect = 1e-16
Identities = 63/230 (27%), Positives = 104/230 (44%)
Frame = +2
Query: 940 PQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVHVVPKKGGMTVVVNEKNELIPT 999
P R+NP +V+K ++ LL+ G I P + G + + + +K+ +
Sbjct: 83 PSYRINPLKLKVLKLQLKDLLEKGFIQPS-----------IYP*GVVVLFLKKKDGFL-- 223
Query: 1000 RTVTGRRMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLDGYSGYNQIAVAPED 1059
RM IDY +LN K +PLP +D++ + L G ++ +D G +Q V ED
Sbjct: 224 ------RMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVIGED 385
Query: 1060 QEKTAFTCPFGVFAYRRMPFGLCGAPATFQRCMLSIFSDMIEKNIEVFMDDFSVFGKSFD 1119
KTAF +G + M FG P F M +F D ++ + VF +D ++ K+ +
Sbjct: 386 VPKTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSKNEN 565
Query: 1120 QCLFHLNAVLKRCTETNLILNWEKCHFMVTEGIVLGHKISSKGIEVDQAK 1169
+ HL LK + L +V G H IS +G++VD +
Sbjct: 566 EHENHLRLALKVLKDIGL-CQISYV*ILVEVGFFSLHVISGEGLKVDSKR 712
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 69.3 bits (168), Expect = 1e-11
Identities = 53/230 (23%), Positives = 95/230 (41%), Gaps = 9/230 (3%)
Frame = +2
Query: 1459 IVWHCHSSAYGGHHSGERTAAKVLQSGFWWPTLFKDCHDFVRRCDNCQ-----RTGSISK 1513
+V H S GH G +++ F+WP + FVR CD C R
Sbjct: 179 LVQESHDSTAAGH-PGRNGTLEIVSRKFFWPGQSQTVRRFVRNCDVCGGIHIWRQAKRGF 355
Query: 1514 RNEMPLTGIIEVEPFDCWGIDFMGPFPPS----SSYLHILVCVDYVTKWVEAIPCVANDS 1569
+P+ + + +DF+ PP+ S YL ++V D ++K V ++
Sbjct: 356 LKPLPVPNRLHSD----LSMDFITSLPPTRGRGSQYLWVIV--DRLSKSVTLEEMDTMEA 517
Query: 1570 KTVVNFLRKNIFTRFGTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVE 1629
+ + G P+ ++SD G ++ F + + ++T YHPQT G E
Sbjct: 518 EACAQRFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLTGVTQLLSTSYHPQTDGGTE 697
Query: 1630 VSNRQLKQILEKTVASSRKDWSRKLDDALWAYRIAFKTHLGLSPYQLVFG 1679
N++++ +L V S+ +W L A R + +G +P+ + G
Sbjct: 698 RWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIGATPFFVEHG 847
>BG582085
Length = 824
Score = 68.6 bits (166), Expect = 2e-11
Identities = 39/77 (50%), Positives = 50/77 (64%), Gaps = 2/77 (2%)
Frame = -3
Query: 1332 KGDSKPRLLRWVLLLQEFDLEIRDKK--GVENVVADHLSRLENNEVTKKEGAIMAEFPDE 1389
KG P LR +LL QEFDLE R ++ G+ ++A L N +VT+ E +I FP+E
Sbjct: 786 KGMQNP*RLRCILLCQEFDLESRQERETGMW*LIA--FPCLRNKQVTQNEISIKERFPNE 613
Query: 1390 QLFAIRERPWFADMANF 1406
QLFAI +RPWFADM NF
Sbjct: 612 QLFAISQRPWFADMTNF 562
Score = 35.0 bits (79), Expect = 0.27
Identities = 22/52 (42%), Positives = 26/52 (49%)
Frame = -2
Query: 1320 FTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIRDKKGVENVVADHLSRLE 1371
F DHA L+Y KGD+KP + I +KG NVVA LS LE
Sbjct: 823 FIDHATLEYC*PKGDAKPVTA*VHSFMPGIRFRI*TRKGDRNVVAYRLSMLE 668
Score = 32.7 bits (73), Expect = 1.3
Identities = 14/24 (58%), Positives = 18/24 (74%)
Frame = -1
Query: 1450 CVAGEEIKNIVWHCHSSAYGGHHS 1473
CV + IK+I+ H HSS+Y GHHS
Sbjct: 530 CVGEKVIKDIL*HHHSSSYMGHHS 459
>BG587150 similar to GP|13592175|gb ppg3 {Leishmania major}, partial (1%)
Length = 754
Score = 68.2 bits (165), Expect = 3e-11
Identities = 50/190 (26%), Positives = 91/190 (47%), Gaps = 5/190 (2%)
Frame = +2
Query: 562 HLAKKKKK-----EEGQFKKFMQLFSQLQVNIPFGDALDQMPVYAKFMKEMLTGRRKPKD 616
+L K+KKK + G +K M+ F + IP ++ + + R+ ++
Sbjct: 95 NLRKRKKKVAKHLKRGANEKEMESFQKRVFMIPLEKPFEEA-YFTHRLWMFFRETRETEE 271
Query: 617 DENISLSENCSAILQRKLPPKLKDPGAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKK 676
D E + +R K D G F I ++ ++ ALC+ GAS+ ++P M
Sbjct: 272 DIRRMFCEAREKMKKRITLKKKSDSGKFAISWTMKGIEFPHALCNTGASVIILPRDMADH 451
Query: 677 LGGGEPKPTRMTLTLADRSISYPFGVLEDVLVKVNDLIFPADFVILDMAEDEDMPLILGR 736
LG + P++ + T D S G++ D+ V++ + + P DF +LD+ + + L+L R
Sbjct: 452 LGL-KVDPSKESFTFVDCSQRSSGGIIRDLEVQIGNALVPVDFHVLDIKINWNSSLLLRR 628
Query: 737 PFLATGRALI 746
FL+TGR +
Sbjct: 629 AFLSTGRGSV 658
>BG580885
Length = 609
Score = 59.3 bits (142), Expect = 1e-08
Identities = 38/131 (29%), Positives = 60/131 (45%), Gaps = 2/131 (1%)
Frame = +3
Query: 111 FDGTGTISPHEHLSHFAETCEFCVPPANVNEDQKKLRLFPFTLIGKAKDW--LLTLPNGT 168
F GT SP HLS F + C N + + + ++FP TL ++ W L P
Sbjct: 54 FRGTPNESPITHLSRFNKVCR----ANNASSVEMQKKIFPVTLEEESALWYDLNIEPYYI 221
Query: 169 IQTWEELELKFLEKYFPMSKYLDKKQEIASFRQGEGESLYDAWERFNLLLKRCPGHEFSE 228
+W+E++L FL+ Y+ + + + E+ QGE E + + R +LKR P H +
Sbjct: 222 SLSWDEIKLSFLQAYYEIEPVEELRSELMGIHQGEKERVRSYFLRLQWILKRWPEHGLED 401
Query: 229 KQYLQFFTEGL 239
F GL
Sbjct: 402 DVIKGVFVNGL 434
>AL367832 weakly similar to GP|14091845|gb Putative retroelement {Oryza
sativa}, partial (2%)
Length = 384
Score = 54.3 bits (129), Expect = 4e-07
Identities = 37/127 (29%), Positives = 59/127 (46%), Gaps = 4/127 (3%)
Frame = -1
Query: 863 ELKELPSH---LKYVFLSKDVSKPAI-ISSTLTPLEEEKLMRVLRENEGALGWKISDLKG 918
E K + H ++ + L + +K I + + L + K+ ++LRE D+ G
Sbjct: 381 ERKAIQPHQEEIELINLGTEENKREIKVGAALEEGVKRKIFQLLREYLDIFACSYEDMPG 202
Query: 919 ISPAYCMHRIHMEAEYKSVVQPQRRLNPTMKEVVKKEVLKLLDAGMIYPISDSSWVSPVH 978
+ P HRI + E V RR +P M +K EV K +DAG + + WV+ +
Sbjct: 201 LDPKIVEHRIPTKPECPPVR*KLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIV 22
Query: 979 VVPKKGG 985
VPKK G
Sbjct: 21 PVPKKDG 1
>AJ497987 weakly similar to GP|9927273|dbj Similar to Arabidopsis thaliana
chromosome II BAC F26H6; putative retroelement pol
polyprotein, partial (1%)
Length = 636
Score = 52.8 bits (125), Expect = 1e-06
Identities = 28/101 (27%), Positives = 56/101 (54%), Gaps = 1/101 (0%)
Frame = -2
Query: 1296 KELLAVIFALEKFRSYLIGSKVIVFTDHAALKYLLTKGDSKPRLLRWVLLLQEFDLEIRD 1355
K A+ +A ++ R Y+I + + +KY+ K R+ RW +LL E+D+E R
Sbjct: 635 KTCCALAWAAKRLRHYMINHTTWLVSKMDPIKYIFEKPALTGRIARWQMLLSEYDIEYRS 456
Query: 1356 KKGVE-NVVADHLSRLENNEVTKKEGAIMAEFPDEQLFAIR 1395
+K ++ +++ADHL+ ++ + I +FPDE++ ++
Sbjct: 455 QKAIKGSILADHLA----HQPLEDYRPIKFDFPDEEIMYLK 345
>BG587166 similar to GP|8778801|gb| T32E20.9 {Arabidopsis thaliana}, partial
(2%)
Length = 756
Score = 51.2 bits (121), Expect = 4e-06
Identities = 29/64 (45%), Positives = 38/64 (59%), Gaps = 1/64 (1%)
Frame = +1
Query: 1763 LFPGKLKSKWSGPFMIESISPYGAVEL-SKPGEPGTFKVNAQRIKPYLGGELPTNKGGVV 1821
LFPGKLKS+ SGPF ++ + PYGA+ L S G F VN QR+K Y+ +
Sbjct: 1 LFPGKLKSR*SGPFKVKEVRPYGAIVLWSTDGR--DFTVNGQRVKLYMATAPEEDGVSTP 174
Query: 1822 LNDP 1825
L+DP
Sbjct: 175 LSDP 186
>CA860311 weakly similar to GP|7289872|gb|A CG17427 gene product {Drosophila
melanogaster}, partial (20%)
Length = 192
Score = 50.4 bits (119), Expect = 6e-06
Identities = 26/61 (42%), Positives = 38/61 (61%), Gaps = 1/61 (1%)
Frame = +1
Query: 1257 ASDYAVGAVLGQR-KNKFFHAIYYASKVLNESQVNYSTTEKELLAVIFALEKFRSYLIGS 1315
A D+ +GA L Q+ + H I YAS++L ++ NY+ E+E LA I+A+ FR YL G
Sbjct: 1 APDWGIGAGLSQKDEENHEHPIAYASRLLTAAERNYTVVERECLAAIWAIRNFRHYLHGP 180
Query: 1316 K 1316
K
Sbjct: 181 K 183
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 686
Score = 50.1 bits (118), Expect = 8e-06
Identities = 22/71 (30%), Positives = 41/71 (56%)
Frame = -2
Query: 1585 GTPRVLISDGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVA 1644
G P +++D G HF +N +++ I+ A+P +PQ++GQ E SN+ + L+K +
Sbjct: 682 GLPYEIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRLD 503
Query: 1645 SSRKDWSRKLD 1655
+ W+ +LD
Sbjct: 502 LKKGCWADELD 470
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 40.4 bits (93), Expect = 0.006
Identities = 15/40 (37%), Positives = 26/40 (64%)
Frame = -1
Query: 1006 RMCIDYRRLNTATRKDHFPLPFMDQMIERLAGQAFYCFLD 1045
RMCIDYR+ N T K+ +PLP +D + +++ ++ +D
Sbjct: 199 RMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF*NID 80
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 37.7 bits (86), Expect = 0.041
Identities = 18/88 (20%), Positives = 42/88 (47%)
Frame = +2
Query: 1593 DGGKHFCNNFLETVLKKYNIKHKVATPYHPQTSGQVEVSNRQLKQILEKTVASSRKDWSR 1652
+G +NF + + K + +++ YHP + GQ E N+ + L + + WS+
Sbjct: 11 EG*SSLYSNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSK 190
Query: 1653 KLDDALWAYRIAFKTHLGLSPYQLVFGK 1680
A + Y ++ ++P++ ++G+
Sbjct: 191 AFPWAEYWYNTSYNISAAMTPFKALYGR 274
>BE997611
Length = 547
Score = 36.2 bits (82), Expect = 0.12
Identities = 21/88 (23%), Positives = 40/88 (44%)
Frame = -2
Query: 559 PYPHLAKKKKKEEGQFKKFMQLFSQLQVNIPFGDALDQMPVYAKFMKEMLTGRRKPKDDE 618
P A ++K+ +F + +VN+P + ++Q+P KF++E+L ++E
Sbjct: 468 PQTEDA*EEKEVPPKFNALHHILKGYKVNMPITETVEQIPCCIKFLQELLKTNANLSEEE 289
Query: 619 NISLSENCSAILQRKLPPKLKDPGAFTI 646
ISLS + + G FT+
Sbjct: 288 FISLSSEFHHTYEVPAVVRFDGEGCFTL 205
>TC80288 weakly similar to PIR|T06029|T06029 hypothetical protein T28I19.100
- Arabidopsis thaliana, partial (14%)
Length = 1460
Score = 35.0 bits (79), Expect = 0.27
Identities = 27/99 (27%), Positives = 45/99 (45%), Gaps = 11/99 (11%)
Frame = +3
Query: 445 EQNKGGFSGNTQENPKNETCNVIELRSK------KVLAPLVPKVPNKVDE-----SVVEV 493
EQ + G GN E + NV E R + K A + + +K +E VE+
Sbjct: 462 EQQQEGEEGNKHETEEESEDNVHERREEQDEEENKHGAEVQEENESKSEEVEDEGGDVEI 641
Query: 494 DENIEDEEVVEKESDQGVVENEKKKKTEGEKSERLIDED 532
DEN ++ + + + VV+ EK K+ EG+ D++
Sbjct: 642 DENDHEKSEADNDREDEVVDEEKDKEEEGDDETENEDKE 758
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.136 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 55,823,219
Number of Sequences: 36976
Number of extensions: 824687
Number of successful extensions: 5287
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 4642
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5178
length of query: 1825
length of database: 9,014,727
effective HSP length: 110
effective length of query: 1715
effective length of database: 4,947,367
effective search space: 8484734405
effective search space used: 8484734405
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146759.6


